Abstract
Scrub typhus is a highly prevalent bacterial infection in India and South Asia that is caused by Orientia tsutsugamushi. The innate immune response to infections is modulated by Toll-like receptors (TLRs) and heat shock proteins (HSPs). This study was done to assess the prevalence and possible association of TLR and HSP polymorphisms in scrub typhus. TLR4 Asp299Gly, TLR4 Thr399Ile, TLR2 Arg753Gln and HSP70-2 A1267G are single-nucleotide polymorphisms (SNPs) that may modulate their activities, and these SNPs were assessed in 137 scrub typhus patients and 134 controls by PCR restriction fragment length polymorphism. We found that the two TLR4 mutations, TLR4 D299G and TLR4T399I, were present in 19.5% and 22% of the study population, respectively, and was in significant linkage disequilibrium with a D’ of 0.8. The TLR2 mutation was found to be rare, whereas the HSP A>G mutation was very common (77.5%). Compared with the controls, the prevalence of heterozygous genotype of the TLR4D299G SNP, but not any of the other SNPs, was significantly higher among scrub typhus patients. Further studies using a larger sample size and more candidate genes may better enable in determining the role of these associations in susceptibility and severity of scrub typhus.
Similar content being viewed by others
Introduction
Scrub typhus is a life-threatening zoonotic bacterial infection prevalent in the Asia-Pacific region.1 The causative organism, Orientia tsutsugamushi is a Gram-negative, obligate intracellular bacterium that infects both nonphagocytic (for example, fibroblasts and endothelial cells) and phagocytic (for example, monocytes/macrophages, polymorphonuclear neutrophils and dendritic cells) cells, and like in other rickettsial diseases, the bacterial infection causes acute vasculitis.2 Macrophages, dendritic cells and neutrophils sense the invading pathogens by the pathogen recognition receptors to activate innate immune response and to promote and orchestrate adaptive immune responses.3 Toll-like receptors (TLRs) are the most important membrane-associated pathogen recognition receptors that recognize pathogen-associated molecular patterns at the cell surface (TLR2, TLR4 and TLR5) or in the intracellular endosomal compartment (TLR3 and TLR7–9).4 There are several reports on the role of TLR2 and TLR4 in the immune response in rickettsial infection,5, 6 but because of the absence of peptidoglycan and lipopolysaccharide (LPS) in its cell wall, the role of TLR2 and TLR4 in the initial interaction between O. tsutsugamushi and innate immune system has been questioned.7, 8 However, these issues are far from clear, and it is not inconceivable that these TLRs are involved in the immune response during O. tsutsugamushi infection, either directly by interacting with structures other than LPS and peptidoglycans in the microbe or indirectly by being activated by, for example, heat shock protein-70 (HSP70) that is released during infection.9
Single-nucleotide polymorphisms (SNPs) have been found in TLR2, TLR4 and HSP70 genes that lead to variation in immune response, and alter susceptibility to various infectious and inflammatory diseases.10, 11 The most studied SNPs for TLRs are the two nonsynonymous, commonly cosegregating mutations, Asp299Gly and Thr399Ile, in TLR4 that alter the ligand binding domain, and the SNP Arg753Glu in TLR2 that causes variation in its function.12, 13 A synonymous polymorphism A1267G in HSP70–2 has been shown to cause reduced production of HSP70 in response to stress.14 Although there are some studies on the associations between these SNPs and the susceptibility to various infections caused by Gram-negative bacteria as well as Mycobacteria,15, 16 the prevalence of these mutations and their association with susceptibility among scrub typhus patients have not been studied. Therefore, this study was designed to examine the prevalence of these mutations among patients with scrub typhus in South India and their possible association with susceptibility.
Materials and methods
This study was conducted at a 2700-bed medical college hospital in South India. Patients above 15 years of age with scrub typhus confirmed by immunoglobulin M enzyme-linked immunosorbent assay and/or PCR for 56 kDa antigen gene of O. tsutsugamushi on the eschar were included as cases. Similar numbers of age and sex-matched, healthy subjects from the same locality of residence who were screened negative for scrub typhus were recruited as controls. Both the patients and controls were natives of Vellore district in Tamil Nadu and belonged to the Tamil-speaking Dravidian ethnic group. The study was approved by the institutional review board and ethics committee. Blood samples from patients and controls were collected after informed and written consent obtained from each participant.
Serological confirmation
Scrub typhus immunoglobulin M enzyme-linked immunosorbent assay was performed on serum samples using the Scrub Typhus Detect (InBios International, Seattle, WA, USA) as per the manufacturer’s instructions. An optical density of >0.5 was considered positive.
PCR analysis for confirmation of O. tsutsugamushi
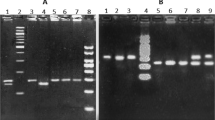
Eschar samples were used for PCR analyses. Bacterial DNA, extracted using a QIAamp DNA mini kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions, was used as the template for the PCR. A standard PCR targeting the 56 kDa protein was carried out as reported previously17 with 1.5 mM MgCl2, 50 mM KCl, 10 mM Tris-HCl (pH 8.3), 200 μM each of dATP, dGTP, dCTP and dTTP, 0.2 μM of primers and 1 U of Taq DNA polymerase (Thermo Scientific, Waltham, MA, USA) in a final volume of 50 μl. The PCR products were electrophoresed in 1.5% agarose gel and the gel containing DNA fragments was stained with 0.5 μg ml−1 of ethidium bromide and visualized by ultraviolet transillumination (Bio-Rad, Hercules, CA, USA).
PCR analyses of restriction fragment length polymorphism (RFLP)
Genomic DNA was isolated from the blood buffy coat by using the commercially available DNA extraction kit (QIAamp from Qiagen). The nonsynonymous SNPs TLR4D299G, TLR4T399I and TLR2R753Q and the synonymous SNP HSP70-2 A1267G were identified using PCR-based RFLP methods. The PCR and RFLP parameters used for the study have been described previously. All the amplifications were carried out in a final volume of 25 μl that included 5 μl of genomic DNA, 1 × PCR Buffer, 20 pmol of primer (10 pmol for TLR4D299G), 1.25 U of Taq polymerase and 0.2 mM dNTP mixture. In the TLR4 gene, the A-to-G mutation in position 896 corresponding to TLR4Asp299Gly SNP introduces a restriction site for the enzyme NcoI (New England Biolabs, Beverly, MA, USA) and the C-to-T mutation at position 1196 corresponding to the TLR4Thr399Ile SNP introduces a restriction site for HinfI, which were used for the RFLP analysis.18, 19 The TLR2Arg753Gln and HSP70-2 polymorphisms were analyzed utilizing the PstI restriction sites introduced at position 2251 of TLR2 gene because of G-to-A mutation and at position 1267 of HSP70–2 gene as a result of A-to-G change.14, 15 All the restriction digestions were carried out at 37 °C overnight, followed by agarose gel electrophoresis and interpretation as described earlier. The primers and restriction enzymes used in the study and the resulting fragment sizes obtained are given in Table 1. The genotyping results were confirmed by sequencing representative samples for each genotype. Sequencing reactions were done using the Big Dye Terminator Cycle Sequencing Kit (Applied Biosystems, Foster City, CA, USA).
Statistical analysis
Statistical analysis was done using R statistical software (www.r-project.org) and the complete genetic analysis was done using ‘SNP assoc’.20 Continuous data were analyzed using mean and s.d. and a P-value of <0.05 was considered significant. The allele and genotype frequencies were determined for each of the SNPs by direct scoring and counting. The risk associated with the SNPs was determined by calculating the odds ratio at 95% confidence intervals. Hardy–Weinberg equilibrium (HWE) calculations were done to compare the observed genotype frequencies with the expected frequencies. P-value and χ2 values were estimated to see that the SNPs were within HWE.
Results
The study population included 145 males (53.5%) and 126 females (46.4%) with a mean age of 41±13 years. Among these, 137 were scrub typhus cases and 134 healthy controls. There was a significant correlation found between the presence of the TLR4Asp299Gly polymorphism in scrub typhus cases compared with controls (P=0.02), whereas no associations were evident with other polymorphisms (Table 2). The overall prevalence of TLR4 D299G, TLR4T399I, TLR2R753Q and HSP-70-2 mutations (heterozygous and homozygous mutant genotypes) in our study was 19.5% (52/271), 22% (59/271), 1.1% (3/368) and 77.5% (203/262), respectively. The genotype and allele frequencies for all the four polymorphisms among cases and controls are shown in Table 2. The frequency of the TLR2R753Q mutation was low and the HSP A>G mutation was very common, with no differences between cases and controls. None of the polymorphisms showed any association with the outcome of the scrub typhus patients.
The TLR4D299G and TLR4T399I were found to lie within HWE (P>0.05) in both cases as well as controls. HWE analysis could not be performed for the TLR2 Arg753Gln control subset because of lack of presence of mutant allele in any control subjects, and the results for the HSP70-2 A1267G cases and controls were not in the HWE (Table 3).
Among the subjects who were heterozygous for TLR4 Asp299Gly, 58% were also found to be heterozygous for TLR4 Thr399Ile, whereas among the wild-type carriers, 86.9% were also wild type for TLR4 Thr399Ile. Of the three homozygous mutants detected in TLR4Asp299Gly, one was homozygous mutant for TLR4 Thr399Ile as well. A significant linkage disequilibrium between TLR4D299G and TLR4 T399I SNP was observed in our population with the linkage disequilibrium values of D= 0.073, D′= 0.82, r2=0.762, χ2=305.4 and P<0.0001, suggesting cosegregation. The haplotype analysis showed that none of the haplotypes had significant association with scrub typhus. Although the haplotype G–C seemed to be associated with scrub typhus with an odds ratio of 2.27 at an α-level of 0.1, with the current sample size, a statistical significance was not observed (95% confidence interval 0.94–5.46; Table 4). None of the other polymorphisms showed any significant correlations with each other.
Discussion
In this study from South India, we found that the two TLR4 mutations, TLR4 D299G and TLR4T399I, were present in 19.5% and 22% of the subjects, respectively, with significant linkage disequilibrium, whereas the TLR2 mutation was found to be very rare and not found among the control population. In contrast to the TLR2 mutation, the HSP A>G mutation was very common (77.5%). Compared with the controls, the prevalence of heterozygous genotype of the TLR4D299G SNP, but not any of the other SNPs, was significantly higher among scrub typhus patients.
The overall prevalence of TLR4D299G polymorphism in our study population is similar to other reports from India,16, 20 and more than that found in Western populations where the incidence is reported to be ∼6 to 14%.21 This polymorphism has been reported to cause LPS hyporesponsiveness, potentially by its ability to impair dimerization of TLR4 and one of its adaptor proteins, MD2, which is of importance for signal transduction.22, 23 This TLR4 mutation has been linked with susceptibility to infections with Gram-negative bacteria and respiratory syncytial virus, as well as reduced risk of noninfectious inflammatory diseases like atherosclerosis or rheumatoid arthritis.11, 22 In India, one study found an association between the severity and bacterial load of pulmonary tuberculosis patients and the TLR4 Asp299Gly mutation, whereas this could not be confirmed in another study.16, 20 In the present study, we, to the best of our knowledge, for the first time report an association between the TLR4D299G polymorphism and the presence of scrub typhus infection. Although the cell wall of O. tsutsugamushi does not contain LPS, one could hypothesize that the hyporesponsive TLR4 mutant, at least through indirect mechanisms, could contribute to an attenuated immune response against the invading microbe, thereby facilitating infection with O. tsutsugamushi.
A D’ value of 0.8 obtained in our study suggests strong linkage disequilibrium between the TLR4D299G and TLR4Thr399Ile mutation, which is in accordance with a previous report from India.16 The overall prevalence of TLR4Thr399Ile mutation, and in particular the A>G mutation in the HSP70–2 gene, was high in our population. Although a study on tuberculosis patients reported a significant association between the presence of wild-type genotype and high bacillary loads compared with the TLR4Thr399Ile mutation,16 and a protective effect of the A>G mutation in the HSP70–2 gene in relation to septic shock has been suggested,10, 14 we could not find any association between these mutations and the presence of scrub typhus infection. A significant deviation from HWE was observed in HSP70-2 A1267G polymorphism in both cases and controls. Although the exact reason for this deviation is not clear, we found no possibility of genotyping errors to explain this trend. There was a higher frequency of HSP 70-2 heterozygotes in both cases and controls in our study, which is similar to earlier reports10, 14 and a significant deviation from HWE for this SNP was reported earlier in a particular Mexican population as well as in black, white and Hispanic females.24, 25
A major limitation of this study is the small sample size of both cases and controls. Further studies looking at a larger study population are required to draw any conclusive correlations between these polymorphisms and susceptibility to scrub typhus. In particular, the low prevalence of TLR2 R753Q mutation in this small sample size makes any conclusion in relation to this mutation impossible.
Conclusion
The present study looked into the possible association of SNPs in three candidate genes in relation to innate immune response with susceptibility to infection with O. tsutsugamushi. A significant correlation was found between the presence of heterozygous TLR4 Asp299Gly and the incidence of scrub typhus. However, larger sample sizes need to be examined to draw positive conclusions of association or dissociation. In relation to our findings on the LPS-hyporesponsive TLR4 mutant, forthcoming studies should evaluate the role of TLR4 activation in the immune response during O. tsutsugamushi infection. Nonetheless, this is a first of its kind study in India on scrub typhus and could provide a new step in deciphering the role of host gene polymorphisms in modulation of immune responses during O. tsutsugamushi infection.
References
Kelly, D. J., Fuerst, P. A., Ching, W. M. & Richards, A. L. Scrub typhus: the geographic distribution of phenotypic and genotypic variants of Orientia tsutsugamushi. Clin. Infect. Dis. 48, S203–S230 (2009).
Jeong, Y. J., Kim, S., Wook, Y. D., Lee, J. W., Kim, K. I. & Lee, S. H. Scrub typhus: clinical, pathologic, and imaging findings. Radiographics 27, 161–172 (2007).
Ting, J. P., Willingham, S. B. & Bergstralh, D. T. NLRs at the intersection of cell death and immunity. Nat. Rev. Immunol. 8, 372–379 (2008).
Blasius, A. L. & Beutler, B. Intracellular toll-like receptors. Immunity 32, 305–315 (2010).
Jordan, J. M., Woods, M. E., Soong, L. & Walker, D. H. Rickettsiae stimulate dendritic cells through toll-like receptor 4, leading to enhanced NK cell activation in vivo. J. Infect. Dis. 199, 236–242 (2009).
Quevedo-Diaz, M. A., Song, C., Xiong, Y., Chen, H., Wahl, L. M., Radulovic, S. et al. Involvement of TLR2 and TLR4 in cell responses to Rickettsia akari. J. Leukoc. Biol. 88, 675–685 (2010).
Mansuelo, P., Vitale, G., Cascio, A., Seidita, A., Pepe, I., Carroccio, A. et al. New insight into immunity and immunopathology of rickettsial diseases. Clin. Dev. Immunol. 2012, 967852 (2011).
Tantibhedhyangkul, W., Amara, A. B., Textoris, J., Gorvel, L., Ghigo, E., Capo, C. et al. Orientia tsutsugamushi, the causative agent of scrub typhus, induces an inflammatory program in human macrophages. Microb. Pathog. 55, 55–63 (2013).
Yonca, B., Michelsen, K. S., Hayrapetian, L., Naiki, Y., Spallek, R., Singh, M. et al. Mycobacterium Tuberculosis heat shock proteins use diverse Toll-like receptor pathways to activate pro-inflammatory signals. J. Biol. Chem. 280, 20961–20967 (2006).
Waterer, G. W., Elbahlawan, L., Quasney, M., Zhang, Q., Kessler, L. & Wuderink, R. G. Heat shock protein 70-2+1267 AA homozygotes have an increased risk of septic shock in adults with community-acquired pneumonia. Crit. Care. Med. 31, 1367–1372 (2003).
Schröder, N. W. & Schumann, R. R. Single nucleotide polymorphisms of Toll-like receptors and susceptibility to infectious disease. Lancet Infect. Dis. 5, 156–164 (2005).
Arbour, N. C., Lorenz, E., Schutte, B. C., Zabner, J., Kline, J. N., Jones, M. et al. TLR4 mutations are associated with endotoxin hyporesponsiveness in humans. Nat. Genet. 25, 187–191 (2000).
Lorenz, E., Mira, J. P., Cornish, K. L., Arbour, N. C. & Schwartz, D. A. A novel polymorphism in the toll-like receptor 2 gene and its potential association with staphylococcal infection. Infect. Immun. 68, 6398–6401 (2000).
Alarcon, G. V., Londono, J. D., Hernandez-Pacheco, G., Gamboa, R., Castillo, E., Pacheco-Tena, C. et al. Heat shock protein 70 gene polymorphisms in Mexican patients with spondyloarthropathies. Ann. Rheum. Dis. 61, 48–51 (2002).
Montes, A. H., Asensi, V., Alvarez, V., Valle, E., Ocaña, M. G., Meana, A. et al. The Toll-like receptor 4(Asp299Gly) polymorphism is a risk factor for Gram-negative and haematogenous osteomyelitis. Clin. Exp. Immunol. 143, 404–413 (2006).
Najmi, N., Kaur, G., Sharma, S. K. & Mehra, N. K. Human Toll-like receptor 4 polymorphisms TLR4 Asp299Gly and Thr399Ile influence susceptibility and severity of pulmonary tuberculosis in the Asian Indian population. Tissue Antigens 76, 102–109 (2010).
Mahajan, S. K., Rolain, J. M., Kashyap, R., Bakshi, D., Sharma, V., Prasher, B. et al. Scrub typhus in Himalayas. Emerg. Infect. Dis. 12, 1590–1592 (2006).
Lee, Y. C., Kim, C., Shim, J. S., Byun, J. Y., Park, M. S., Cha, C. I. et al. Toll-like receptors 2 and 4 and their mutations in patients with otitis media and middle ear effusion. Clin. Exp. Otorhinolaryngol. 1, 189–195 (2008).
Lorenz, E., Frees, K. L. & Schwartz, D. A. Determination of the TLR4 genotype using allele-specific PCR. Biotechniques 31, 22–24 (2001).
Gonzalez, J. R., Armengol, L., Sole, X., Guino, E., Mercader, J. M., Estivill, X. et al. SNPassoc: an R package to perform whole genome association studies. Bioinformatics 23, 644–645 (2007).
Selvaraj, P., Harishankar, M., Singh, B., Jawahar, M. S. & Banurekha, V. V. Toll-like receptor and TIRAP gene polymorphisms in pulmonary tuberculosis patients of South India. Tuberculosis 90, 306–310 (2010).
Ferwerda, B., Alonso, S., Giamarellos-Bourboulis, E. J., Mouktaroudi, M., Izagirre, N., Syafruddin, D. et al. TLR4 Polymorphisms, infectious diseases, and evolutionary pressure during migration of modern humans. Proc. Natl Acad. Sci. USA 104, 16645–16650 (2007).
Figueroa, L., Xiong, Y., Song, C., Piao, W., Vogel, S. N. & Medvedev, A. E. Asp299Gly polymorphism alters TLR4 signaling by interfering with recruitment of MyD88 and TRIF. J Immunol. 188, 4506–4515 (2012).
Zuniga, J., Vargas-Alarcon, G., Osnaya, N., Martinez-Tripp, S., Rodriguez-Reyna, T. S., Hernandez-Martinez, B. et al. Heat-shock protein (HSP70-2) allelic frequencies in three distinct Mexican populations. Genes Immun. 1, 66–68 (1999).
Nguyen, D. P., Genc, M., Vardhana, S., Babula, O., Onderdonk, A. & Witkin, S. S. Ethnic differences of polymorphisms in cytokine and innate immune system genes in pregnant women. Obstet. Gynecol. 104, 293–300 (2004).
Acknowledgements
This work was supported by Grants from University of Oslo. We thank Mr S Suresh Kumar for the technical assistance provided for the work.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Rights and permissions
About this article
Cite this article
Janardhanan, J., Joseph Martin, S., Astrup, E. et al. Single-nucleotide polymorphisms in Toll-like receptor (TLR)-2, TLR4 and heat shock protein 70 genes and susceptibility to scrub typhus. J Hum Genet 58, 707–710 (2013). https://doi.org/10.1038/jhg.2013.89
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/jhg.2013.89