Abstract
The aim of this study was to investigate association between copy number variation of the defensin beta 4 gene (DEFB4) and susceptibility to cervical cancer in a population at high risk of persistent oncogenic human papillomavirus (HPV) infection. The study subjects comprised 204 women with cervical cancer, a population having a high risk of persistent oncogenic HPV infection (cervical cancer group), and 200 healthy women from the general population (control group). Copy number variation of DEFB4 in each test sample was determined by relative quantitation using the comparative CT (ΔΔCT) method. Differences between the two groups were evaluated. The median DEFB4 copy number in the cervical cancer group was four and in the control group was five (P=2.77e–4, t-test). The odds ratio of cervical cancer in individuals with four DEFB4 copies or less was higher (odds ratio 2.02; 95% confidence interval odds ratio 1.36–3.02), compared with that in individuals with five or more copies (odds ratio 0.49; 95% confidence interval odds ratio 0.33–0.74). We found copy number variation of DEFB4 was a host genetic factor conferring susceptibility to cervical cancer. A lower DEFB4 copy number was associated with susceptibility to cervical cancer.
Similar content being viewed by others
Introduction
In humans, β-defensins are a family of small cationic antimicrobial peptides that exert an important first-line antimicrobial activity against a broad spectrum of bacteria, fungi and viruses.1 The β-defensins are expressed in a variety of epithelia.2 In particular, the defensin beta 4 gene (DEFB4; MIM 602215) gene is expressed in the non-pregnant female reproductive tract, including in the uterus, cervix and vagina.3 A cluster of antimicrobial β-defensin genes on 8p23.1, including DEFB4, SPAG11, DEFB103, DEFB104, DEFB105, DEFB106 and DEFB107, exhibits copy number variation (CNV) in the population.4 Individuals have 2–12 copies of this cluster per diploid genome. Levels of DEFB4 mRNA are significantly correlated with DEFB4 copy number.5 Higher copy numbers of DEFB4, result in significantly upregulated transcription after induction by tumor necrosis factor-α, which results in better antimicrobial activity in vitro.6 Conversely, a lower DEFB4 copy number may lead to decreased antimicrobial activity. The median copy number of DEFB4 per diploid genome was significantly lower in human immunodeficiency virus-positive children (3.54) compared with that in human immunodeficiency virus-exposed uninfected children (4.49).7 CNV of DEFB4 is associated with susceptibility to human immunodeficiency virus infection.8 Therefore, CNV of DEFB4 on 8p23.1 may be associated with susceptibility to infectious disease.
Cervical cancer is the third most common cancer in women worldwide. Persistent infection with oncogenic human papillomavirus (HPV), including types HPV-16, HPV-18, HPV-31, HPV-33, HPV-45, HPV-52 and HPV-58, is recognized as a major risk factor for cervical cancer,9 but is not sufficient on its own for cancer development. Only a small proportion of women with persistent infection of oncogenic HPV over many years develop carcinoma in situ and invasive cervical cancer. Genital infections with HPV are very common, and these infections are transmitted by sexual contact.10 The HPV life cycle is intimately linked to the differentiation program of the epithelium targeted by HPV. HPVs are thought to infect the basal cells of the epithelium through microtraumas, and viral genomes are maintained as episomes in the basal layer, with viral gene expression being tightly controlled as the infected cells move toward the epithelial surface.11 However, HPV infections in most cases disappear naturally in a relatively short period, and pose little risk of developing disease.12, 13, 14 Additional cofactors; for example, host genetic factors and/or environmental factors, may be involved in cervical carcinogenesis. HPV cofactors have been identified, including oral contraceptive use and smoking.15 Possible factors in persistent infection could be related to the host, such as the host immune reaction against an oncogenic HPV infection. For example, several reports suggest that genetic variations in human leukocyte antigen class II regions influence the risk of cervical cancer by altering the efficiency of the immune response to HPV infections.16, 17, 18, 19, 20 However, we do not fully understand the pathological mechanism that results in HPV infection developing into invasive cervical cancer.
In this study, we hypothesized that CNV in the DEFB4 gene may be associated with susceptibility to cervical cancer. To investigate our hypothesis, we compared DEFB4 CNV in women with cervical cancer, a population with a high risk for persistent oncogenic HPV infections, with that in healthy women from the general population.
Materials and methods
Patient backgrounds
The study populations comprised 204 women with cervical cancer, a population having a high risk of persistent oncogenic HPV infection (cervical cancer group), and 200 healthy women from the general population (control group). All of these women attended the Department of Obstetrics and Gynecology at Nagasaki University Hospital or its satellite hospitals from August 2006 to December 2009. All samples were obtained after receiving written informed consent, and the study protocol was approved by the Institutional Review Board for Ethical, Legal and Social Issues of Nagasaki University. All cases of cervical cancer were diagnosed as squamous cell carcinoma by histopathological examinations, whereas healthy women had no abnormal cytological findings in pap smear tests of the uterine cervix. There were no significant differences between the cervical cancer group and the control group for the following factors: age, parity, body mass index, smoking habit and use of oral contraceptives (P>0.05, Table 1).
Genomic DNA samples
Genomic DNA was extracted from peripheral whole blood using a DNA Purification Kit (Qiagen, Hilden, Germany) according to the manufacturer's protocol. Genomic DNA samples were prepared at 5 ng μl–1 using a NanoDrop ND-1000 spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA).
Analysis of genomic DNA samples with known copy numbers of DEFB4 by TaqMan real-time PCR
The albumin gene (ALB) served as a two copies per diploid genome reference gene. The method of quantitative real-time PCR using LightCycler Relative Quantification Software 1.0 (Roche, Mannheim, Germany) requires an appropriate calibrator. Genomic DNA sample, C0913, with three copies of DEFB4 per diploid genome and two copies of ALB per diploid genome, was purchased from the European Collection of Cell Cultures (Wiltshire, UK) as the calibrator.21 Real-time PCR was performed using the LightCycler480 (Roche). The set of primers and the hybridization probe sequence for ALB (GenBank accession no. M12523) were: forward primer: 5′-TGGAAAATGATGAGATGCCTG-3′, reverse primer: 5′-CATAGTTTTTGCAAACATCC-3′, FAM reporter: 5′-CTTTCAACAAAATCAGCAGCTAATGAAGGC-3′, and those for DEFB4 exon 2 (NM_04942) were: forward primer: 5′-TTTTGGTGGTATAGG-3′, reverse primer: 5′-TCTAGGGCAAAAGACTG-3′, FAM reporter: 5′-ATGGCTCCACTCTTAAGGCAGGTAACAG-3′. All PCR reaction mixtures were prepared automatically using QIAgility (Qiagen). Real-time PCR reactions (10 μl) were carried out in triplicate with 10 ng of unknown genomic DNA or calibrator DNA (three copies of DEFB4 and two copies of ALB per diploid genome), 2 × Quanti-Tect-Multiplex-PCR NoROX buffer (Qiagen), 0.125 μM of each primer and 0.0625 μM of each TaqMan probe. Thermal cycling was initiated with a 2-min incubation at 50 °C, followed by a first denaturation step of 15 min at 95 °C, and then by 40 cycles of 15 s at 95 °C and 1 min at 60 °C, followed by one cycle at 5 °C for 6 min. The number of copies of DEFB4 in each test sample was determined by relative quantitation using the comparative CT (ΔΔCT) method.22 This method measured the cycle threshold (CT) difference (ΔCT) between DEFB4 as a target and ALB as a reference and then compared the ΔCT values of test samples to the calibrator sample known to have three copies of the target sequence. All samples were analyzed in triplicate. Compared with the genomic DNA calibrator, with 3 DEFB4 copies per diploid genome, genomic DNA samples, with 2, 4, 5, 6, 7 and 8 copies of DEFB4 and 2 copies of ALB per diploid genome, were identified from blood samples of healthy women.
Determination of DEFB4 CNV in genomic DNA samples
To create standard curves for each known DEFB4 copy number (2, 3, 4, 5, 6, 7 and 8 copies) and for 2 copies of ALB in the diploid genome, we selected individuals from the general population known to have each known copy number and compared these with C0913, which has three DEFB4 copies per diploid genome. In all, 20, 10, 5 and 2.5 ng of genomic DNA samples for each known copy number individual were prepared as a two-step dilution series. Then, for each dilution, the DEFB4 and the ALB genes were amplified in a single assay to acquire the relative quantification standard curves for both genes (Figure 1). The genomic DNA samples with unknown copies of DEFB4 were amplified in each run in parallel with the samples with known copy numbers of DEFB4. In addition, each PCR run was referenced to the control. A no-template control (negative control) was also included in each assay.
Standard curve for copy number quantification of defensin beta 4 gene (DEFB4). The standard curves of each known copy number of DEFB4 (2, 3, 4, 5, 6, 7 and 8 copies) and 2 copies of albumin gene (ALB) in the diploid genome are shown as black lines and ±2 s.d. areas. The vertical axis indicates DEFB4 copy number, whereas the horizontal axis shows log2 genomic DNA concentrations. Each plot reflects DEFB4 assay copy numbers versus log2 (ng) genomic DNA concentration. The DEFB4 copy number for genomic DNA samples with unknown copy number is classified into each copy number area.
Statistical analysis
Differences between the two groups were evaluated with Student's t-test. Odds ratios are estimated with 95% confidence intervals using logistic regression. Statistical analyses were performed with SPSS software version 19 (IBM, Armonk, NY, USA). Significant differences were defined as P-values of <0.05.
Results
Characteristics of the two groups are described in Table 1. The cervical cancer group and the control group were similar with regard to age, parity, body mass index, smoking and oral contraceptive use.
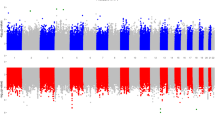
DEFB4 CNV is described as the mean (s.d.) number of copies, and its distribution in cervical cancer and control groups is shown in Figure 2. CNV of DEFB4 in the control group was 4.69 (1.45) copies, whereas that in the cervical cancer group was 4.25 (1.25) copies, showing that the DEFB4 copy number in the cervical cancer group was significantly lower than in the control group (P=2.77e–4, t-test).
Distribution of defensin beta 4 gene (DEFB4) copy number variations (CNVs) in 204 women with cervical cancer and in 200 healthy women. Mean (s.d.) DEFB4 CNV was 4.25 (1.25) copies in the cervical cancer group, and 4.69 (1.45) copies in healthy women. There was a significant difference in DEFB4 CNV between the two groups (P=2.77e–4, t-test). The black bar indicates the distribution of DEFB4 CNVs in the cervical cancer group, while the white bar indicates the distribution in healthy women.
The odds ratio of cervical cancer in individuals with four copies or less of DEFB4 was relatively higher (odds ratio 2.02; 95% confidence interval odds ratio 1.36–3.02), compared with that in individuals with five copies or more (odds ratio 0.49; 95% confidence interval odds ratio 0.33–0.74). Therefore, both values indicated a two-tailed significant difference from an odds ratio of 1.00 at the 5% level.
Discussion
To our knowledge, this is the first study to investigate the association of DEFB4 copy number with susceptibility to cervical cancer in a population at high risk of persistent oncogenic HPV infection. We found that a lower DEFB4 gene copy number is associated with susceptibility to cervical cancer.
Cervical cancer develops via two critical transition steps: persistent HPV infection and progression to cervical intraepithelial neoplasia grade 3/invasive cancer (invasion through the basement membrane of the epithelium). Thus, HPV infection alone is not sufficient to cause cervical intraepithelial neoplasia grade 3/cervical cancer, and host genetic factors are very likely to contribute to cervical cancer pathogenesis.23 A number of studies have evaluated the association of host genetic variations with cervical cancer and have implicated a role for human leukocyte antigens class II.16, 17, 18, 19, 20 Many case–control studies have consistently reported a protective effect of human leukocyte antigen DRB1*13 against cervical cancer. Several class II alleles, such as DRB1*1501, DRB1*1302, DQB1*0602 and DQB1*03 are associated with an increased risk of cervical cancer, although the strength of the associations vary across racial and ethnic groups. In the population-based cohort studies of women in Guanacaste, Costa Rica, single-nucleotide polymorphisms in the Fanconi anemia complementation group A (FANCA) gene were associated with cervical cancer susceptibility. Single-nucleotide polymorphisms in the interferon regulatory factor 3 (IRF3) gene, which is an innate immune gene, and in the 2′,5′ oligoadenylate synthetase 3 (OAS3), sulfatase 1 (SULF1), deoxyuridine triphosphate (DUT) and general transcription factor IIH, polypeptide 3 (GTF2H4) genes were associated with HPV persistence. However, single-nucleotide polymorphisms in the interferon gamma (IFNG), epidermal dysplasia verruciformis (EV)-associated EVER1/EVER2, peroxiredoxin 3 (PRDX3) and ribosomal protein S19 (RPS19) genes were associated with progression to cervical intraepithelial neoplasia grade 3/cancer.24, 25, 26 In this study, a lower DEFB4 copy number was associated with susceptibility to cervical cancer, although the copy number at which DEFB4 exerts its protective effect is not fully understood. The copy number of DEFB4 affects its level of expression.5 Thus, lower DEFB4 CNV may increase susceptibility to cervical cancer because of persistent oncogenic HPV infection via a dosage effect. However, DEFB4 only displayed mild anti-HPV activity in vitro.27 Another study has shown substantial genetic heterogeneity in the human β-defensin gene cluster that may be a result of selection.28 Hence, more widespread associations may be needed to determine whether such genetic heterogeneity accounts for some of the geographical or ethnic variations in different populations. And, as a cluster of β-defensin genes at 8p23.1 includes six genes (DEFB4, DEFB103, DEFB104, DEFB105, DEFB106 and DEFB107) within 100 kb region, there is a possibility that the copy number of DEFB4 must closely link to that of other defensin genes. Therefore, to obtain more valuable information regarding the relation between CNVs of DEFB4 and susceptibility to cervical cancer, the expression level of DEFB4 should be analyzed in cervical tissues with low copy number and those with high copy number. Also, genome-wide association studies or exome sequencing analysis of cervical cancer should shed additional light on host genetic factors involved in cervical cancer. This new information may clarify the interplay between viral and host genetic factors in HPV persistence and the risk of progression to cervical intraepithelial neoplasia grade 3/cancer.
In conclusion, DEFB4 CNV was observed in both cervical cancer and healthy control groups, and a lower DEFB4 copy number may be associated with susceptibility to cervical cancer, which is a population at high risk of persistent oncogenic HPV infection. We found CNV of DEFB4 to be an additional host genetic susceptibility factor of cervical cancer. Further studies are required to determine the reproducibility of our findings in larger patient groups and different study populations.
Accession codes
References
Ganz, T. Defensins: antimicrobial peptides of innate immunity. Nat. Rev. Immunol. 3, 710–720 (2003).
Schutte, B. C. & McCray, P. B. [Beta]-defensins in lung host defense. Annu. Rev. Physiol. 64, 709–748 (2002).
Horne, A. W., Stock, S. J. & King, A. E. Innate immunity and disorders of the female reproductive tract. Reproduction 135, 739–749 (2008).
Hollox, E. J., Armour, J. A. L. & Barber, J. C. K. Extensive normal copy number variation of a beta-defensin antimicrobial-gene cluster. Am. J. Hum. Genet. 73, 591–600 (2003).
Groth, M., Wiegand, C., Szafranski, K., Huse, K., Kramer, M., Rosenstile, P. et al. Both copy number and sequence variations affect expression of human DEFB4. Genes Immun. 11, 458–466 (2010).
Janssens, W., Nuytten, H., Dupont, L. J., Van Eldere, J., Vermeire, S., Lambrechts, D. et al. Genomic copy number determines functional expression of {beta}-defensin 2 in airway epithelial cells and associates with chronic obstructive pulmonary disease. Am. J. Respir. Crit. Care Med. 182, 163–169 (2010).
Milanese, M., Segat, L., Arraes, L. C., Garzino-Demo, A. & Crovella, S. Copy number variation of defensin genes and HIV infection in Brazilian children. J. Acquir. Immune Defic. Syndr. 50, 331–333 (2009).
Sun, L., Finnegan, C. M., Kish-Catalone, T., Blumenthal, R., Garzino-Demo, P., La Terra Maggiore, G. M. et al. Human beta-defensins suppress human immunodeficiency virus infection: potential role in mucosal protection. J. Virol. 79, 14318–14329 (2005).
Schiffman, M., Castle, P. E., Jeronimo, J., Rodriguez, A. C. & Wacholder, S. Human papillomavirus and cervical cancer. Lancet 370, 890–907 (2007).
Shimada, T., Miyashita, M., Miura, S., Nakayama, D., Miura, K., Fukuda, M. et al. Genital human papilloma virus infection in mentally-institutionalized virgins. Gynecol. Oncol. 106, 488–489 (2007).
Doorbar, J. Molecular biology of human papillomavirus infection and cervical cancer. Clin. Sci. 110, 525–541 (2006).
Moscicki, A. B., Palefsky, J., Smith, G., Siboshski, S. & Schoolnik, G. Variability of human papillomavirus DNA testing in a longitudinal cohort of young women. Obstet. Gynecol. 82 (Pt 1), 578–585 (1993).
Woodman, C. B., Collins, S., Winter, H., Bailey, A., Ellis, J., Prior, P. et al. Natural history of cervical human papillomavirus infection in young women: a longitudinal cohort study. Lancet 357, 1831–1836 (2001).
Ho, G. Y., Bierman, R., Beardsley, L., Chang, C. J. & Burk, R. D. Natural history of cervicovaginal papillomavirus infection in young women. N. Engl. J. Med. 338, 423–428 (1998).
Castellsague, X. & Munoz, N. Cofactors in human papillomavirus carcinogenesis–role of parity, oral contraceptives, and tobacco smoking. J. Natl. Cancer Inst. Monogr. 31, 20–28 (2003).
Apple, R. J., Erlich, H. A., Klitz, W., Manos, M. M., Becker, T. M. & Wheeler, C. M. HLA DR-DQ associations with cervical carcinoma show papillomavirus-type specificity. Nat. Genet. 6, 157–162 (1994).
Beskow, A. H., Josefsson, A. M. & Gyllensten, U. B. HLA class II alleles associated with infection by HPV16 in cervical cancer in situ. Int. J. Cancer 93, 817–822 (2001).
Hildesheim, A. & Wang, S. S. Host and viral genetics and risk of cervical cancer: a review. Virus Res. 89, 229–240 (2002).
Wang, S. S. & Hildesheim, A. Viral and host factors in human papillomavirus persistence and progression. J. Natl. Cancer Inst. Monogr. 31, 35–40 (2003).
Matsumoto, K., Maeda, H., Oki, A., Takatsuka, N., Yasugi, T., Furuta, R. et al. HLA class II DRB1*1302 allele protects against progression to cervical intraepithelial neoplasia grade 3: a multicenter prospective cohort study. Int. J. Gynecol. Cancer 22, 471–478 (2012).
Perne, A., Zhang, X., Lehmann, L., Groth, M., Stuber, F. & Book, M. Comparison of multiplex ligation-dependent probe amplification and real-time PCR accuracy for gene copy number quantification using the beta-defensin locus. Biotechniques 47, 1023–1028 (2009).
Chen, Q., Book, M., Fang, X., Hoeft, A. & Stuber, F. Screening of copy number polymorphisms in human beta-defensin genes using modified real-time quantitative PCR. J. Immunol. Methods 308, 231–240 (2006).
Magnusson, P. K., Lichtenstein, P. & Gyllensten, U. B. Heritability of cervical tumours. Int. J. Cancer 88, 698–701 (2000).
Wang, S. S., Bratti, M. C., Rodriguez, A. C., Herrero, R., Burk, R. D., Porras, C. et al. Common variants in immune and DNA repair genes and risk for human papillomavirus persistence and progression to cervical cancer. J. Infect. Dis. 199, 20–30 (2009).
Wang, S. S., Gonzalez, P., Yu, K., Porras, C., Li, Q., Safaeian, M. et al. Common genetic variants and risk for HPV persistence and progression to cervical cancer. PLoS One 5, e8667 (2010).
Safaeian, M., Hildesheim, A., Gonzalez, P., Yu, K., Porras, C., Li, Q. et al. Single nucleotide polymorphisms in the PRDX3 and RPS19 and risk of HPV persistence and cervical precancer/cancer. PLoS One 7, e33619 (2012).
Buck, C. B., Day, P. M., Thompson, C. D., Lubkowski, J., Lu, W., Lowy, D. R. et al. Human alpha-defensins block papillomavirus infection. Proc. Natl Acad. Sci. 103, 1516–1521 (2006).
Hardwick, R. J., Machado, L. R., Zuccherato, L. W., Antolinos, S., Xue, Y., Shawa, N. et al. A worldwide analysis of beta-defensin copy number variation suggests recent selection of a high-expressing DEFB103 gene copy in East Asia. Hum. Mutat. 32, 743–750 (2011).
Acknowledgements
We thank Miss Yasuko Noguchi for her technical assistance. This work was supported by the Japan Society for the Promotion of Science KAKENHI grant numbers nos. 23592406, 24791712 and 22591827.
Author contributions: All authors must see and approve the final version of the manuscript. Conceived and designed the experiment: KM, KiY and HM. Performed the experiments: SA, KM, AK, HM, SM and KY. Analyzed the data: SA, KM, YH and KS. Contributed materials and analysis tools: AH, OJ, KS and AY. Wrote the paper: SA and KM.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Abe, S., Miura, K., Kinoshita, A. et al. Copy number variation of the antimicrobial-gene, defensin beta 4, is associated with susceptibility to cervical cancer. J Hum Genet 58, 250–253 (2013). https://doi.org/10.1038/jhg.2013.7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/jhg.2013.7
Keywords
This article is cited by
-
Convergent evolution of defensin sequence, structure and function
Cellular and Molecular Life Sciences (2017)
-
Low DEFB4 Copy Number and High Systemic hBD-2 and IL-22 Levels Are Associated with Dermatophytosis
Journal of Investigative Dermatology (2015)
-
A novel method for sex determination by detecting the number of X chromosomes
International Journal of Legal Medicine (2015)
-
Single human papillomavirus 16 or 52 infection and later cytological findings in Japanese women with NILM or ASC-US
Journal of Human Genetics (2014)