Abstract
We ascertained two large Pakistani consanguineous families (PKDF231 and PKDF608) segregating profound hearing loss, vestibular dysfunction, and retinitis pigmentosa; the defining features of Usher syndrome type 1 (USH1). To date, seven USH1 loci have been reported. Here, we map a novel locus, USH1K, on chromosome 10p11.21–q21.1. In family PKDF231, we performed a genome-wide linkage screen and found a region of homozygosity shared among the affected individuals at chromosome 10p11.21–q21.1. Meiotic recombination events in family PKDF231 define a critical interval of 11.74 cM (20.20 Mb) bounded by markers D10S1780 (63.83 cM) and D10S546 (75.57 cM). Affected individuals of family PKDF608 were also homozygous for chromosome 10p11.21–q21.1-linked STR markers. Of the 85 genes within the linkage interval, PCDH15, GJD4, FZD4, RET and LRRC18 were sequenced in both families, but no potential pathogenic mutation was identified. The USH1K locus overlaps the non-syndromic deafness locus DFNB33 raising the possibility that the two disorders may be caused by allelic mutations.
Similar content being viewed by others
Introduction
Usher syndrome (USH)-causing genes have provided unexpected insights into developmental and biochemical processes shared by the eye and ear.1, 2 USH is a neurosensory disorder affecting both hearing and vision in humans.3, 4 A molecular diagnosis study suggested a frequency of 1/6000 individuals afflicted with USH in the United States.5 Among the three defined clinical subtypes, USH type I (USH1) is the most genetically heterogeneous. To date, seven loci for USH1, three loci for USH2 and one locus for USH3 and genes for nine of them have been described.6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17 All of the USH proteins, including myosin VIIa (MIM 276903), cadherin 23 (MIM 605516), protocadherin 15 (MIM 605514), harmonin (MIM 605242), SANS (MIM 607696), usherin (MIM 608400), GPR98 (MIM 602851), whirlin (MIM 607928) and clarin-1 (MIM 606397), are thought to interact to form a large macromolecular complex,1, 18 which is essential for auditory and visual functions. Furthermore, growing evidence suggests that USH proteins are localized in inner ear stereocilia bundles and are part of the mechano-transduction machinery, and in the retina, largely within the molecular network tethering cilium to photoreceptor cells.1, 11, 19, 20, 21, 22, 23, 24, 25, 26 However, the molecular identities of many essential components of these structures are unknown, precluding our understanding of the precise mechanisms of human hearing and vision.
Discovering new USH genes requires the investigation of large human families segregating for USH, followed by genetic mapping and positional identification of causal mutations. Consanguineous families, plentiful in the Pakistani population, represent a rich resource for gene discovery research. Here we report two consanguineous Pakistani families in which the USH1 phenotype is linked to a novel USH1 locus, USH1K, on chromosome 10p11.21–q21.1.
Materials and methods
Subject enrollment
This study was approved by IRB Committee at the Cincinnati Children’s Hospital Research Foundation, USA (2009-0684; 2010-0291), the IRB at the National Centre of Excellence in Molecular Biology, Lahore, Pakistan (FWA00001758) and the Combined Neuroscience IRB at the National Institutes of Health, USA (OH-93-N-016). Written informed consent was obtained from all adult subjects and parents of minor subjects under the age of 18 years. Subjects in this study were ascertained from the Punjab province of Pakistan. Probands were initially identified at schools for the deaf in Punjab, Pakistan. The National Centre of Excellence in Molecular Biology, Lahore, Pakistan enrolled the families in this study and performed clinical evaluations in collaboration with National Institutes of Health. Genotyping and sequencing of the DNA samples from the participating individuals was carried out at the Cincinnati Children’s Hospital Research Foundation, USA.
Clinical evaluation
We performed medical history interviews to find obvious syndromic and environmental causes of hearing loss. For some of the affected individuals, a physical examination was performed to detect signs and symptoms of other disorders such as Waardenburg or Pendred syndromes. Affected subjects underwent a general otological examination, including otoscopic examination and audiometry. Hearing was evaluated in some affected and unaffected subjects by pure-tone air and bone-conduction audiometry with or without tympanometry. No air-bone gaps were observed in any tested individuals. Vestibular function was assessed by tandem gait and Romberg testing. Funduscopic examinations were performed by an ophthalmologist to confirm the absence or presence of retinitis pigmentosa.
DNA isolation, genotyping and linkage analysis
Genomic DNA was extracted from peripheral blood samples using a standard protocol.27 We performed a genome-wide scan in family PKDF231 for homozygosity among offspring of consanguineous marriages using 388 STR markers (v2.5 ABI Prism Linkage Mapping Set, Applied Biosystems, Foster City, CA, USA) and an ABI 3730 Genetic Analyzer. Alleles were assigned using Gene Mapper software (Applied Biosystems). Linkage in family PKDF608 was identified by screening with STR markers linked to chromosome 10p11.21–q21.1.
LOD score calculations
Marker order and map distances are from the Marshfield genetic map (http://research.marshfieldclinic.org/). Two-point LOD scores were calculated with Superlink online version 1.5 (http://bioinfo.cs.technion.ac.il/superlink-online/). We assumed a recessive mode of inheritance, with full penetrance of USH in homozyotes and no phenocopies. The disease allele frequency was set at 0.001 with equal meiotic recombination frequencies for males and females. Short tandem repeat allele frequencies were defined by genotype analyses of 100 unaffected Pakistani individuals.
Candidate genes
We identified candidate USH1K genes on the UCSC Human Genome Browser (http://genome.ucsc.edu/). PCDH15, GJD4, FZD4, RET and LRRC18 genes were sequenced in both PKDF231 and PKDF608 families using the primers flanking all of the exonic and adjacent intronic sequences. PCR, sequencing conditions and mutation analysis procedures were performed essentially as described.28, 29
Results
Clinical description
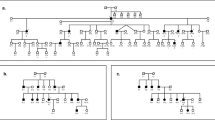
At the time of examination, the ages of the affected individuals in family PKDF231 (Figure 1a) ranged from 12 to 26 years, whereas the ages of the affected individuals of family PKDF608 (Figure 1a) ranged from 18 to 45 years. All affected individuals in both families displayed congenital, bilateral and profound sensorineural hearing loss (Figure 1b). Affected individuals in both families had delayed onset of independent ambulation, consistent with vestibular dysfunction, which was further confirmed by tandem gait ability and by using the Romberg test. Funduscopic examination of two older affected individuals of both families PKDF231 (V:5 (24 years old) and V:7 (26 years old); Figure 1a) and PKDF608 (V:20 (45 years old) and V:21 (18 years old); Figure 1a) revealed signs of retinitis pigmentosa along with narrowing of retinal blood vessel, bone spicules and waxy appearance of optic disk. The severity of retinitis pigmentosa was directly related to the age of the patient and ranged from mild to severe loss of vision.
USH1K families PKDF231 and PKDF608 and their representative audiograms. (a) Chromosome 10 haplotypes in family PKDF231. Filled symbols represent affected individuals. The USH1J-linked haplotype is boxed. The STR markers and physical map positions in megabases (Mb, February 2009 human reference sequence GRCh37, hg19) are shown on the left of the pedigree. Haplotype analysis of PKDF231 shows a linkage region of 20.20 Mb delimited by markers D10S1780 (35.89 Mb) and D10S546 (56.09 Mb). Affected individuals V:2, V:3, V:4, V:5 and V:7 provided distal meiotic breakpoint at D10S546 (56.09 Mb), whereas the unaffected individual V:6 provided the proximal recombination at D10S1780 (35.89 Mb). In family PKDF608, affected individual V:18 provided the proximal meiotic breakpoint at marker D10S1780 (35.89 Mb). The distal breakpoint at marker D10S1652 (64.31 Mb) was provided by affected individual V:29 (data not shown). (b) Pure tone air and bone conduction thresholds for family PKDF231 V:7 (26-year-old male), and family PKDF608 V:20 (45-year-old male) revealed profound, bilateral, sensorineural hearing loss. Right ear air conduction: O; Left ear air conduction: × ; Right ear bone conduction: >; Left ear bone conduction: <;↓ indicates the threshold level beyond the measurable range.
Linkage mapping
We undertook a genome-wide linkage analysis in family PKDF231. It initially showed suggestive evidence of linkage only to markers on chromosome 10p11.21–q21.1. Affected individuals were homozygous for markers in this interval, whereas unaffected obligate carriers were heterozygous (Figure 1a). Additional markers were genotyped and haplotype analysis revealed a 11.74-cM interval of homozygosity delimited by markers D10S1780 (63.83 cM) and D10S546 (75.57 cM; Figure 2). A maximum two-point LOD score (Zmax) of 3.82 at recombination fraction θ=0 was obtained for the marker D10S539 (Table 1). Chromosome 10p11.21–q21.1-linked STR markers were then used to screen additional families segregating USH or isolated recessive deafness. One additional family, PKDF608, was found to be segregating USH1 linked to markers in this region (Figure 1a). A maximum two-point LOD score (Zmax) of 3.22 at recombination fraction θ=0 was obtained for the marker D10S539 (Table 1).
USH1K linkage intervals in families PKDF231 and PKDF608 on human chromosome 10p11.21–q21.1. STR markers are represented by filled circles. The sex-averaged recombination positions in cM are indicated for STR markers. Candidate genes in the USH1K interval were identified from the UCSC Human Genome Browser February 2009 assembly (http://genome.ucsc.edu/). Candidate genes expressed in the inner ear are underlined. Previously reported deafness locus DFNB33 interval and USH1F/DFNB23-causing gene PCDH15 is also shown.
Mutant alleles of PCDH15 are responsible for USH1F/DFNB23 phenotype in humans.7, 28, 29, 30 In all, 24 exons of PCDH15 are present within the distal breakpoint and therefore we considered it a candidate for the USH1 phenotype. Full sequencing of PCDH15 in the genomic DNA from two affected individuals from each family along with a normal hearing sibling did not reveal any functional mutations. Therefore, the HUGO nomenclature committee assigned the USH1K designation for the locus defined by families PKDF231 and PKDF608. Linkage interval of the USH1K locus is approximately 20.20 Mb delimited by markers D10S1780 and D10S546 and harbors 85 candidate genes (Figure 2). The USH1K critical interval overlaps DFNB33 (MIM 607239), a locus for non-syndromic recessively inherited hearing loss that was previously mapped between markers D10S193 and D10S1784.31 We next considered the possibility that mutations of a single gene might underlie both USH1K and DFNB33. If so, the mutated gene is located between markers D10S1780 and D10S1784, which spans 19.12 Mb (UCSC human genome browser, Figure 2). On this assumption, we examined the overlapping linkage interval of USH1K and DFNB33 and found the same 85 genes (Figure 2). Analyses of data reported in a massively parallel signature sequencing libraries of mRNA from inner ear tissues32 and the SHIELD database (https://shield.hms.harvard.edu/) reveal that 53 of the 85 genes are expressed in the inner ear (Figure 2). The coding exons and flanking intronic sequence of additional four candidate genes GJD4, FZD8, RET and LRRC18 were sequenced in two affected individuals from each of the two USH1K families analyzed in this study, and no pathogenic sequence variants were found.
Discussion
Haplotype analysis of two families revealed a 11.74-cM region of homozygosity for USH1K on chromosome 10p11.21–q21.1. Families PKDF231 and PKDF608 each have unique haplotypes across this region, and may segregate different mutant USH1 alleles. The USH1K locus overlaps the DFNB33 locus on chromosome 10 and these two hearing disorders may be due to allelic mutations. Mutant alleles of four of the known USH1 genes, MYO7A, USH1C, CDH23 and PCDH15 are responsible for both non-syndromic hearing loss and USH.2, 7, 8, 9, 10, 13, 14, 28, 33, 34, 35 However, it is also plausible that these two loci are non-allelic.
In addition to USH1K and DFNB33 loci, human chromosome 10q also harbors two other loci for USH type 1 and nonsyndromic hearing loss, USH1D/DFNB12 and USH1F/DFNB23.6, 7, 9, 10, 33 The USH1D/DFNB12 locus is approximately 17.10 Mb telomeric to USH1K locus, whereas part of the PCDH15 gene, responsible for USH1F/DFNB23, lies within the distal boundary of USH1K (Figure 2). Sequencing of all the known coding and non-coding exons and 100 bp flanking exon–intron junctions of PCDH15 in both USH1K families did not reveal any pathogenic mutation. Although it is possible that PCDH15 may harbor cryptic mutations in cochlear-specific regulatory regions leading to USH1 in these two families, however, statistical analysis did not provide significant evidence of linkage of the USH1 phenotype segregating in USH1K family PKDF231 to PCDH15 intronic STR marker (D10S546; Table 1). There have been precedents for two closely associated or partially overlapping deafness loci in human, for example, DFNB36 and DFNB96, DFNB3 and DFNB85, DFNB35 and 14q23.1–q31.1 loci.36, 37, 38, 39
In the linkage interval common to USH1K and DFNB33, there appears to be 85 known genes, out of which 53 are expressed in the inner ear. The candidate deafness genes in the critical USH1K/DFNB33 interval are GJD4, FZD4, RET and LRRC18 (Figure 2). GJD4 (MIM 611922) encodes the gap junction protein connexin 40.1. Mutations in several different connexin sub-units have been identified in individuals suffering with either non-syndromic or syndromic deafness.40 FZD4 (MIM 604579) encodes a seven-transmembrane domain protein that belongs to frizzled receptor gene family. FZD4 has a central role in the inner ear vascular development through Wnt signaling pathway.41 RET (MIM 164761), a member of cadherin superfamily, encodes a receptor tyrosine kinase.42 Mutations in RET also cause Hirschsprung disease (MIM 142623), in which hearing loss is sometimes detected.43, 44 LRRC18 encodes a leucine-rich repeat containing protein member 18. Mutations in a different family member, LRTOMT (also known as LRRC51; MIM 612414), have been implicated in non-syndromic hearing loss.45, 46, 47 However, sequencing of these four candidate genes in both USH1K families did not reveal any potential pathogenic variant. All of the 85 candidate genes and conserved sequences in the USH1J/DFNB33 interval will now need to be screened for mutant alleles. Rather than continuing hierarchical sequencing of candidate genes based on function or expression, future studies will employ massively parallel sequencing of genomic DNA from the affected individuals of these families enriched for the entire USH1J critical interval.
USH mutations are estimated to be responsible for more than 50% of deaf-blindness, 8–33% of patients thought to have isolated RP, and 3–6% of patients are thought to have isolated deafness.48, 49, 50 Effects of hearing loss on quality of life include difficulty in understanding speech and social isolation. In routine life, people with blindness or deafness are often able to compensate with other senses, whereas individuals with USH are deficient in both senses and thus suffer exacerbated quality-of-life effects. Ultimately, by limiting one’s ability to communicate and interact, hearing and vision impairments impact cognitive, emotional and social development, making the development of intervention strategies a clinically significant long-term goal of the current research. Mapping of USH1K is another step towards understanding the molecular mechanisms resulting in USH.
References
Kremer, H., van Wijk, E., Marker, T., Wolfrum, U., Roepman, R. Usher syndrome: molecular links of pathogenesis, proteins and pathways. Hum. Mol. Genet. 15 Spec No 2 R262–R270 (2006).
Petit, C. Usher syndrome: from genetics to pathogenesis. Annu. Rev. Genomics Hum. Genet. 2, 271–297 (2001).
Smith, R. J., Berlin, C. I., Hejtmancik, J. F., Keats, B. J., Kimberling, W. J., Lewis, R. A. et al. Clinical diagnosis of the Usher syndromes. Usher Syndrome Consortium. Am. J. Med. Genet. 50, 32–38 (1994).
Ahmed, Z. M., Riazuddin, S., Wilcox, E. R. The molecular genetics of Usher syndrome. Clin. Genet. 63, 431–444 (2003).
Kimberling, W. J., Hildebrand, M. S., Shearer, A. E., Jensen, M. L., Halder, J. A., Trzupek, K. et al. Frequency of Usher syndrome in two pediatric populations: Implications for genetic screening of deaf and hard of hearing children. Genet. Med. 12, 512–516 (2010).
Ahmed, Z. M., Riazuddin, S., Bernstein, S. L., Ahmed, Z., Khan, S., Griffith, A. J. et al. Mutations of the protocadherin gene PCDH15 cause Usher syndrome type 1F. Am. J. Hum. Genet. 69, 25–34 (2001).
Alagramam, K. N., Yuan, H., Kuehn, M. H., Murcia, C. L., Wayne, S., Srisailpathy, C. R. et al. Mutations in the novel protocadherin PCDH15 cause Usher syndrome type 1F. Hum. Mol. Genet. 10, 1709–1718 (2001).
Bitner-Glindzicz, M., Lindley, K. J., Rutland, P., Blaydon, D., Smith, V. V., Milla, P. J. et al. A recessive contiguous gene deletion causing infantile hyperinsulinism, enteropathy and deafness identifies the Usher type 1C gene. Nat. Genet. 26, 56–60 (2000).
Bolz, H., von Brederlow, B., Ramírez, A., Bryda, E. C., Kutsche, K., Nothwang, H. G. et al. Mutation of CDH23, encoding a new member of the cadherin gene family, causes Usher syndrome type 1D. Nat. Genet. 27, 108–112 (2001).
Bork, J. M., Peters, L. M., Riazuddin, S., Bernstein, S. L., Ahmed, Z. M., Ness, S. L. et al. Usher syndrome 1D and nonsyndromic autosomal recessive deafness DFNB12 are caused by allelic mutations of the novel cadherin-like gene CDH23. Am. J. Hum. Genet. 68, 26–37 (2001).
Ebermann, I., Scholl, H. P., Charbel Issa, P., Becirovic, E., Lamprecht, J., Jurklies, B. et al. A novel gene for Usher syndrome type 2: mutations in the long isoform of whirlin are associated with retinitis pigmentosa and sensorineural hearing loss. Hum. Genet. 121, 203–211 (2007).
Joensuu, T., Hämäläinen, R., Yuan, B., Johnson, C., Tegelberg, S., Gasparini, P. et al. Mutations in a novel gene with transmembrane domains underlie Usher syndrome type 3. Am. J. Hum. Genet. 69, 673–684 (2001).
Verpy, E., Leibovici, M., Zwaenepoel, I., Liu, X. Z., Gal, A., Salem, N. et al. A defect in harmonin, a PDZ domain-containing protein expressed in the inner ear sensory hair cells, underlies Usher syndrome type 1C. Nat. Genet. 26, 51–55 (2000).
Weil, D., Blanchard, S., Kaplan, J., Guilford, P., Gibson, F., Walsh, J. et al. Defective myosin VIIA gene responsible for Usher syndrome type 1B. Nature 374, 60–61 (1995).
Weil, D., El-Amraoui, A., Masmoudi, S., Mustapha, M., Kikkawa, Y., Lainé, S. et al. Usher syndrome type I G (USH1G) is caused by mutations in the gene encoding SANS, a protein that associates with the USH1C protein, harmonin. Hum. Mol. Genet. 12, 463–471 (2003).
Weston, M. D., Eudy, J. D., Fujita, S., Yao, S., Usami, S., Cremers, C. et al. Genomic structure and identification of novel mutations in usherin, the gene responsible for Usher syndrome type IIa. Am. J. Hum. Genet. 66, 1199–1210 (2000).
Weston, M. D., Luijendijk, M. W., Humphrey, K. D., Moller, C., Kimberling, W. J. Mutations in the VLGR1 gene implicate G-protein signaling in the pathogenesis of Usher syndrome type II. Am. J. Hum. Genet. 74, 357–366 (2004).
El-Amraoui, A., Petit, C. Usher I syndrome: unravelling the mechanisms that underlie the cohesion of the growing hair bundle in inner ear sensory cells. J. Cell Sci. 118, 4593–4603 (2005).
Ahmed, Z. M., Goodyear, R., Riazuddin, S., Lagziel, A., Legan, P. K., Behra, M. et al. The tip-link antigen, a protein associated with the transduction complex of sensory hair cells, is protocadherin-15. J. Neurosci. 26, 7022–7034 (2006).
Kazmierczak, P., Sakaguchi, H., Tokita, J., Wilson-Kubalek, E. M., Milligan, R. A., Müller, U. et al. Cadherin 23 and protocadherin 15 interact to form tip-link filaments in sensory hair cells. Nature 449, 87–91 (2007).
Maerker, T., van Wijk, E., Overlack, N., Kersten, F. F., McGee, J., Goldmann, T. et al. A novel Usher protein network at the periciliary reloading point between molecular transport machineries in vertebrate photoreceptor cells. Hum. Mol. Genet. 17, 71–86 (2008).
Overlack, N., Maerker, T., Latz, M., Nagel-Wolfrum, K., Wolfrum, U. SANS (USH1G) expression in developing and mature mammalian retina. Vision Res. 48, 400–412 (2008).
Lagziel, A., Overlack, N., Bernstein, S. L., Morell, R. J., Wolfrum, U., Friedman, T. B. Expression of cadherin 23 isoforms is not conserved: implications for a mouse model of Usher syndrome type 1D. Mol. Vis. 15, 1843–1857 (2009).
Goodyear, R. J., Forge, A., Legan, P. K., Richardson, G. P. Asymmetric distribution of cadherin 23 and protocadherin 15 in the kinocilial links of avian sensory hair cells. J. Comp. Neurol. 518, 4288–4297 (2010).
Caberlotto, E., Michel, V., Foucher, I., Bahloul, A., Goodyear, R. J., Pepermans, E. et al. Usher type 1G protein sans is a critical component of the tip-link complex, a structure controlling actin polymerization in stereocilia. Proc. Natl Acad. Sci. USA 108, 5825–5830 (2011).
Grati, M., Kachar, B. Myosin VIIa and sans localization at stereocilia upper tip-link density implicates these Usher syndrome proteins in mechanotransduction. Proc. Natl Acad. Sci. USA 108, 11476–11481 (2011).
Grimberg, J., Nawoschik, S., Belluscio, L., McKee, R., Turck, A., Eisenberg, A. A simple and efficient non-organic procedure for the isolation of genomic DNA from blood. Nucleic Acids Res. 17, 8390 (1989).
Ahmed, Z. M., Riazuddin, S., Bernstein, S. L., Ahmed, Z., Khan, S., Griffith, A. J. et al. Mutations of the protocadherin gene PCDH15 cause Usher syndrome type 1F. Am. J. Hum. Genet. 69, 25–34 (2001).
Ahmed, Z. M., Riazuddin, S., Aye, S., Ali, R. A., Venselaar, H., Anwar, S. et al. Gene structure and mutant alleles of PCDH15: nonsyndromic deafness DFNB23 and type 1 Usher syndrome. Hum. Genet. 124, 215–223 (2008).
Ben-Yosef, T., Ness, S. L., Madeo, A. C., Bar-Lev, A., Wolfman, J. H., Ahmed, Z. M. et al. A mutation of PCDH15 among Ashkenazi Jews with the type 1 Usher syndrome. N. Engl. J. Med. 348, 1664–1670 (2003).
Belguith, H., Masmoudi, S., Medlej-Hashim, M., Chouery, E., Weil, D., Ayadi, H. et al. Re-assigning the DFNB33 locus to chromosome 10p11.23-q21.1. Eur. J. Hum. Genet. 17, 122–124 (2009).
Peters, L. M., Belyantseva, I. A., Lagziel, A., Battey, J. F., Friedman, T. B., Morell, R. J. Signatures from tissue-specific MPSS libraries identify transcripts preferentially expressed in the mouse inner ear. Genomics 89, 197–206 (2007).
Ahmed, Z. M., Riazuddin, S., Ahmad, J., Bernstein, S. L., Guo, Y., Sabar, M. F. et al. PCDH15 is expressed in the neurosensory epithelium of the eye and ear and mutant alleles are responsible for both USH1F and DFNB23. Hum. Mol. Genet. 12, 3215–3223 (2003).
Ahmed, Z. M., Smith, T. N., Riazuddin, S., Makishima, T., Ghosh, M., Bokhari, S. et al. Nonsyndromic recessive deafness DFNB18 and Usher syndrome type IC are allelic mutations of USHIC. Hum. Genet. 110, 527–531 (2002).
Riazuddin, S., Nazli, S., Ahmed, Z. M., Yang, Y., Zulfiqar, F., Shaikh, R. S. et al. Mutation spectrum of MYO7A and evaluation of a novel nonsyndromic deafness DFNB2 allele with residual function. Hum. Mutat. 29, 502–511 (2008).
Ansar, M., Lee, K., Naqvi, S. K., Andrade, P. B., Basit, S., Santos-Cortez, R. L. et al. A new autosomal recessive nonsyndromic hearing impairment locus DFNB96 on chromosome 1p36.31-p36.13. J. Hum. Genet. 56, 866–868 (2011).
Naz, S., Griffith, A. J., Riazuddin, S., Hampton, L. L., Battey, J. F., Khan, S. N. et al. Mutations of ESPN cause autosomal recessive deafness and vestibular dysfunction. J. Med. Genet. 41, 591–595 (2004).
Shahin, H., Walsh, T., Rayyan, A. A., Lee, M. K., Higgins, J., Dickel, D. et al. Five novel loci for inherited hearing loss mapped by SNP-based homozygosity profiles in Palestinian families. Eur. J. Hum. Genet. 18, 407–413 (2010).
Friedman, T. B., Liang, Y., Weber, J. L., Hinnant, J. T., Barber, T. D., Winata, S. et al. A gene for congenital, recessive deafness DFNB3 maps to the pericentromeric region of chromosome 17. Nat. Genet. 9, 86–91 (1995).
Scott, C. A., Kelsell, D. P. Key functions for gap junctions in skin and hearing. Biochem. J. 438, 245–254 (2011).
Xu, Q., Wang, Y., Dabdoub, A., Smallwood, P. M., Williams, J., Woods, C. et al. Vascular development in the retina and inner ear: control by Norrin and Frizzled-4, a high-affinity ligand-receptor pair. Cell 116, 883–895 (2004).
Traugott, A. L., Moley, J. F. The RET protooncogene. Cancer Treat. Res. 153, 303–319 (2010).
Skinner, R., Irvine, D. Hirschsprung’s disease and congenital deafness. J. Med. Genet. 10, 337–339 (1973).
Liang, J. C., Juarez, C. P., Goldberg, M. F. Bilateral bicolored irides with Hirschsprung's disease. A neural crest syndrome. Arch. Ophthalmol. 101, 69–73 (1983).
Ahmed, Z. M., Masmoudi, S., Kalay, E., Belyantseva, I. A., Mosrati, M. A., Collin, R. W. et al. Mutations of LRTOMT, a fusion gene with alternative reading frames, cause nonsyndromic deafness in humans. Nat. Genet. 40, 1335–1340 (2008).
Vanwesemael, M., Schrauwen, I., Ceuppens, R., Alasti, F., Jorssen, E., Farrokhi, E. et al. A 1 bp deletion in the dual reading frame deafness gene LRTOMT causes a frameshift from the first into the second reading frame. Am. J. Med. Genet. A 155A, 2021–2023 (2011).
Du, X., Schwander, M., Moresco, E. M., Viviani, P., Haller, C., Hildebrand, M. S. et al. A catechol-O-methyltransferase that is essential for auditory function in mice and humans. Proc. Natl Acad. Sci. USA 105, 14609–14614 (2008).
Boughman, J. A., Vernon, M., Shaver, K. A. Usher syndrome: definition and estimate of prevalence from two high-risk populations. J. Chronic Dis. 36, 595–603 (1983).
Brownstein, Z., Ben-Yosef, T., Dagan, O., Frydman, M., Abeliovich, D., Sagi, M. et al. The R245X mutation of PCDH15 in Ashkenazi Jewish children diagnosed with nonsyndromic hearing loss foreshadows retinitis pigmentosa. Pediatr. Res. 55, 995–1000 (2004).
Vernon, M. Usher’s syndrome--deafness and progressive blindness. Clinical cases, prevention, theory and literature survey. J. Chronic Dis. 22, 133–151 (1969).
Acknowledgements
We are grateful to the families who made this research possible. These families were ascertained using the intramural funds from NIDCD DC000039-15 to Thomas B Friedman. We thank Khitab Gul, R Amjad Ali and Hashim Raza for technical assistance and Tom Friedman for his suggestions regarding this manuscript. ZMA is a recipient of an RPB Career Development Award. This work was supported by the Higher Education Commission and Ministry of Science and Technology, Islamabad, Pakistan, to Shaikh Riazuddin the International Center for Genetic Engineering and Biotechnology, Trieste, Italy under project CRP/PAK08-01 contract no. 08/009 to Shaikh Riazuddin Cincinnati Children’s Hospital Research Foundation (CCHMC) Intramural Research Funds, to SR and ZMA; Action on Hearing Loss grant to SR; National Institute on Deafness and Other Communication Disorders (NIDCD/NIH) research grants R00 DC009287 to ZMA and R01 DC011803 to SR.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Rights and permissions
About this article
Cite this article
Jaworek, T., Bhatti, R., Latief, N. et al. USH1K, a novel locus for type I Usher syndrome, maps to chromosome 10p11.21–q21.1. J Hum Genet 57, 633–637 (2012). https://doi.org/10.1038/jhg.2012.79
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/jhg.2012.79
Keywords
This article is cited by
-
Novel and recurrent CIB2 variants, associated with nonsyndromic deafness, do not affect calcium buffering and localization in hair cells
European Journal of Human Genetics (2016)