Abstract
Mucopolysaccharidosis type VI (MPS VI) is a genetic disorder caused by a deficiency of arylsulfatase B (ARSB). In our previous study, we investigated the structural changes in ARSB caused by amino acid substitutions associated with MPS VI, and revealed that such structural changes in ARSB were correlated with the clinical phenotypes. To the best of our knowledge, there is no database containing the structures of mutant ARSBs. Here, we built a database of clinical phenotypes, genotypes and structures of mutant ARSBs (http://mps6-database.org). This database can be accessed via the Internet, and is user friendly being equipped with powerful computational tools. This database will be useful for a better understanding of MPS VI.
Similar content being viewed by others
Main
Arylsulfatase B (ARSB, N-acetylgalactosamine-4–sulfatase, EC 3.1.6.12) catalyzes the hydrolysis of the sulfate moiety of glycosaminoglycan (GAG) dermatan sulfate.1, 2 A deficiency of ARSB, which is caused by a mutation in the ARSB gene located on chromosome 5 (5q13-5q14), results in the accumulation of the substrate in lysosomes of various types of tissues, leading to an autosomal recessive lysosomal storage disorder, mucopolysaccharidosis type VI (MPS VI; Maroteaux–Lamy syndrome; MIM#253200). MPS VI exhibits a broad spectrum of clinical phenotypes, from severe to attenuated forms, and patients with this disease exhibit growth retardation, a short stature, coarse faces, skeletal deformities, stiff joints, corneal clouding, respiratory difficulty, hepatosplenomegaly and cardiac abnormalities.
So far, a large number of ARSB gene mutations, predominantly missense ones, causing MPS VI have been identified,1, 2 and their complexity makes it difficult to understand the disease. To elucidate the mechanism underlying the disease, three-dimensional (3D) structural analysis of ARSB has been performed. For example, Garrido et al.3 visualized the locations of mutations in the ARSB structure using 3D visualization software. Furthermore, our group revealed that the structural changes in ARSB caused by amino acid substitutions were correlated with the clinical phenotypes;4 that is, a large structural change in ARSB or a structural change in the core region of ARSB tends to cause a severe form, whereas a small structural change in ARSB or a structural change on the surface of ARSB tends to cause an attenuated form. This suggests that information on structural changes in ARSB will facilitate our understanding of the disease.
In this study, we built a database of clinical phenotypes, genotypes and structures of mutant ARSBs. The information on the ARSB gene mutations was mainly obtained from the HGMD database (http://www.hgmd.org/),5 and structural models of mutant ARSBs were built according to the method described previously,4 using the crystal structure of human ARSB as a template (Protein Data Bank (PDB) code: 1FSU).6 All researchers and clinicians can use this database (http://mps6-database.org) for free. To use all the functions of this database, JavaScript and Java Runtime Environment must be plugged in.
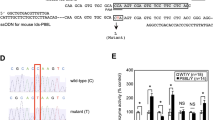
A total of 96 unique ARSB mutations (81 missense mutations and 15 nonsense) have been incorporated into the database so far. However, the structural data on the template ARSB from the PDB do not provide us with information about the locations of two specific missense mutations in the molecule, so mutant ARSB models for the 79 missense ones were built. To the best of our knowledge, this is the first database of the 3D structures of mutant ARSBs, and it contains: (i) comprehensive information on the gene mutations associated with MPS VI (data structure and basic statistics are presented in Supplementary data S1 and S2, respectively), (ii) tools for 3D structure visualization and (iii) tools for searching for ARSB gene mutations. Several tools have been installed within the database to enhance its scope. A text search tool is provided for searching selected fields of the database. A control table option is incorporated for an intensive search. Using this option, users can search for MPS VI gene mutations associated with specific phenotypes. Figure 1 shows the page of the list of MPS VI gene mutations. This database also allows users to observe the 3D structures of the mutant proteins using Jmol (http://www.jmol.org), which is an open-source Java viewer for chemical structures. The database provides users with many options for visualizing the structures of mutant ARSBs. Figure 2 presents a page concerning the mutant ARSB structure with a G308R amino acid substitution, as an example.
The page of the list of MPS VI gene mutations. The ‘phenotype’ is determined basically according to the original papers as described in Saito et al.4
Color imaging of a mutant ARSB protein with a G308R amino acid substitution. Each atom of the molecule is colored according to the distance between the atom in the mutant and the corresponding atom in the wild-type structure. The colors of the atoms show the distances, as follows: blue<0.15 Å, 0.15 Å⩽cyan<0.30 Å, 0.30 Å⩽green<0.45 Å, 0.45 Å⩽yellow<0.60 Å, 0.60 Å⩽orange<0.75 Å and red⩾0.75 Å. The colors of the atoms show the distances, as follows: blue<0.15 Å, 0.15 Å ⩽cyan <0.30 Å, 0.30 Å ⩽green<0.45 Å, 0.45 Å⩽yellow<0.60 Å, 0.60 Å⩽orange< 0.75 Å, and red ⩾0.75 Å.
In conclusion, we built a database for MPS VI. This database will help users to understand MPS VI.
References
Neufield, E. F. & Muenzer, J. in The Metabolic & Molecular Bases of Inherited Disease (eds Scriver, C. R., Beaudet, A. L., Sly, W. S. & Valle, D.) 3421–3452 (McGraw-Hill, New York, 2001).
Valayannopoulos, V., Nicely, H., Harmatz, P. & Turbeville, S. Mucopolysaccharidosis VI. Orphanet J. Rare Dis. 5, 5 (2010).
Garrido, E., Cormand, B., Hopwood, J. J., Chabás, A., Grinberg, D. & Vilageliu, L. Maroteaux-Lamy syndrome: functional characterization of pathogenic mutations and polymorphisms in the arylsulfatase B gene. Mol. Genet. Metab. 94, 305–312 (2008).
Saito, S., Ohno, K., Sugawara, K. & Sakuraba, H. Structural and clinical implications of amino acid substitutions in N-acetylgalactosamine-4-sulfatase: Insight into mucopolysaccharidosis type VI. Mol. Genet. Metab. 93, 419–425 (2008).
Cooper, D. N. & Krawczak, M. Human gene mutation database. Hum. Genet. 98, 629 (1996).
Bond, C. S., Clements, P. R., Ashby, S. J., Collyer, C. A., Harrop, S. J., Hopwood, J. J. et al. Structure of a human lysosomal sulfatase. Structure 5, 277–289 (1997).
Acknowledgements
This work was supported by the Japan Society for the Promotion of Science (IDs: 21390314 and 23659527, HS); the Program for Promotion of Fundamental Studies in Health Sciences of the National Institute of Biomedical Innovation (ID: 09-15, HS); the Program for Research on Intractable Disease of Health and Labor Science Research Grants (HS); the JAPS Asia/Africa Scientific Platform Program (HS); and the High-Tech Research Center Project of the Ministry of Education, Culture, Sports, Science and Technology of Japan (S0801043, HS).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies the paper on Journal of Human Genetics website
Supplementary information
Rights and permissions
About this article
Cite this article
Saito, S., Ohno, K., Sekijima, M. et al. Database of the clinical phenotypes, genotypes and mutant arylsulfatase B structures in mucopolysaccharidosis type VI. J Hum Genet 57, 280–282 (2012). https://doi.org/10.1038/jhg.2012.6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/jhg.2012.6
Keywords
This article is cited by
-
The clinical and genetic Spectrum of Maroteaux-Lamy syndrome (Mucopolysaccharidosis VI) in the Eastern Province of Saudi Arabia
Journal of Community Genetics (2018)
-
Mucopolysaccharidosis VI in cats – clarification regarding genetic testing
BMC Veterinary Research (2016)
-
Factors and processes modulating phenotypes in neuronopathic lysosomal storage diseases
Metabolic Brain Disease (2014)
-
Mucopolysaccharidosis type VI phenotypes-genotypes and antibody response to galsulfase
Orphanet Journal of Rare Diseases (2013)