Abstract
Sotos syndrome is an overgrowth syndrome with characteristic facial dysmorphism, variable severity of learning disabilities and macrocephaly with overgrowth. Haploinsufficiency of the nuclear receptor SET domain-containing protein 1 (NSD1) gene located on 5q35 has been implicated as the cause of Sotos syndrome. This study was performed to investigate the mutation spectrum of NSD1 abnormalities and meaningful genotype−phenotype correlations in Korean patients with Sotos syndrome. Eighteen unrelated Korean patients with Sotos syndrome were enrolled for clinical and molecular analyses. Cytogenetic studies were performed to confirm 5q35 microdeletion, and NSD1 sequencing analysis was performed to identify intragenic mutations. NSD1 abnormalities were identified in 15 (83%) patients. Among them, eight patients (53%) had 5q35 microdeletions and the other seven patients (47%) had seven different NSD1 intragenic mutations including four novel mutations. The mutation spectrum of Korean patients with Sotos syndrome was similar to that of previous studies for Japanese patients. Height was significantly shorter and age of walking alone was significantly older in the microdeletion group compared with those in the intragenic mutation group. No significant differences were observed for other clinical characteristics between the microdeletion and intragenic mutation groups. Further studies with a larger number of patients will be necessary to draw conclusive genotype−phenotype correlations.
Similar content being viewed by others
Introduction
Sotos syndrome (OMIM 117550) is an autosomal dominantly inherited congenital malformation syndrome belonging to the overgrowth condition group. Clinical diagnostic criteria have been defined as Sotos et al.1, 2, 3 firstly described the syndrome in 1964. The diagnostic criteria are based on the presence of a characteristic facial appearance, learning disabilities and macrocephaly with prenatal and postnatal overgrowth.4 The typical facial appearance includes a prominent broad forehead, sparse fronto−temporal hair, down-slanting palpebral fissures and a pointed chin. In addition, various abnormalities, including maternal pre-eclampsia, neonatal jaundice, neonatal hypotonia, poor feeding in infancy, seizures, scoliosis, advanced bone age, joint laxity, and cardiac, renal and brain malformations can accompany the other features.5
Since 2002, Sotos syndrome has been associated with haploinsufficiency of the nuclear receptor SET domain-containing protein 1 (NSD1) gene located on 5q35.6 NSD1 consists of 23 exons and encodes multiple functional domains, including the SU(VAR)3-9, E(Z), tirthorax (SET), SET-associated domains, which mediate histone methyltransferase activity, five plant homeo-domains implicated in chromatin regulation and two proline−tryptophan−tryptophan−proline domains that may mediate protein interactions.3, 7, 8, 9 NSD1 is expressed in several tissues including the brain, kidney, skeletal muscle, spleen and thymus.10 Although an exact role of the NSD1 protein has not been identified, the presence of two different ligand-binding domains suggests that NSD1 enables the regulation of both negative and positive transcription.11
Several reports have demonstrated NSD1 abnormalities in patients with Sotos syndrome.12, 13, 14, 15 NSD1 abnormalities include microdeletion of 5q35, encompassing the entire NSD1 deletion (deletion type) and mutations within the NSD1 gene (intragenic mutation type). To date, over 300 different mutations associated with Sotos syndrome have been identified, including gross deletions, small indels, point mutations and splice site mutations (HGMD, http://www.hgmd.org/).
Notably, 5q35 microdeletions are more frequently (∼50%) found in Japanese patients with Sotos syndrome,6, 16, 17 whereas 5q35 microdeletions are uncommon (<15%) in patients outside of Japan.3, 18 Moreover, NSD1 abnormalities have been delineated in up to 90% of non-Japanese patients with Sotos syndrome, whereas ∼30% of Japanese patients do not have NSD1 abnormalities.19
However, the exact mechanism of such an inter-racial difference has not been clearly explained to date. Patients with a microdeletion tend to have certain congenital heart and/or urogenital anomalies, more severe mental retardation and shorter stature than those with NSD1 intragenic mutations.20 In this study, we delineated the clinical characteristics and mutational spectrum of NSD1 in 18 unrelated Korean patients with Sotos syndrome.
Materials and methods
Patients
Eighteen unrelated Korean patients (6 females and 12 males; age range, 1.3–14.5 years) were enrolled. All patients were clinically diagnosed with Sotos syndrome by clinical diagnostic criteria,1, 2, 3 and showed a normal karyotype. The patients were evaluated at the Department of Medical Genetics, Ajou University Hospital and the Department of Pediatrics, Seoul National University Hospital, Korea. The study protocols were reviewed and approved by the Institutional Review Boards of these two hospitals, and written informed consent was obtained from all subjects or from their parents. Clinical assessment was performed by review of medical records. Physical growth was evaluated by Korean reference data21 and expressed as a standard deviation score (SDS).
NSD1 analyses
Fluorescence in situ hybridization (FISH) analysis for 5q35 microdeletion was performed first to investigate NSD1 abnormalities, considering the genetic spectrum of Japanese patients with Sotos syndrome. If microdeletion was found in FISH analysis, additional array comparative genomic hybridization (aCGH) was performed to measure the deletion size. If the FISH analysis revealed no deletion, NSD1 sequencing analysis was performed successively.
Fluorescence in situ hybridization
The FISH study for delineation of the 5q35 microdeletion was performed on metaphase spreads of cultured peripheral leukocytes stimulated with phytohemagglutinin using an NSD1 probe (Vysis LSI NSD1, Abbott Laboratories, Chicago, IL, USA) on 5q35.
Array comparative genomic hybridization
Applied microarray platform was the NimbleGen, CGX-3 135 K Whole-Genome Array (Roche Diagnostics, Mannheim, Germany). Patients’ genomic and reference DNA were labeled with Cy3 and Cy5, respectively. Equal amounts of labeled test and reference DNA were used for hybridization on one array, which was then processed according to the manufacturer's instructions. Array images were analyzed by the Genoglyphix software (Signature Genomics Laboratories, Spokane, WA, USA).
DNA sequencing
Blood specimens were collected from patients and their parents. Genomic DNA was isolated from peripheral blood leukocytes, according to the manufacturer’s instructions using a Wizard Genomic DNA Purification kit (Promega, Madison, WI, USA). All coding exons and exon−intron boundaries of the NSD1 gene were amplified by PCR using appropriate primers designed by the authors (available on request) and a thermal cycler (Applied Biosystems, Foster City, CA, USA). Direct sequencing was performed using a BigDye Terminator Cycle Sequencing Ready Reaction kit (Applied Biosystems), and the analysis was conducted using an ABI3130xl Genetic Analyzer (Applied Biosystems).
Statistical analyses
All continuous variables are expressed as medians and ranges. Statistical significance (P<0.05) was determined by the Mann–Whitney test for continuous variables and the Fisher’s exact test for nominal variables using PASW version 18.0 (SPSS, Chicago, IL, USA).
Results
Clinical findings
The clinical features of the patients are listed in Table 1. All patients showed characteristic facial dysmorphism and developmental delay. The median age for walking alone was 24 months (range, 14–40 months) and for speaking a meaningful word was 24 months (range, 12–72 months). Median height SDS and weight SDS were 1.4 (range, −0.47–3.0) and 1.2 (range, −1.3–2.6), respectively. Median head circumference SDS was 2.6 (range, 1.2–3.9). Thirteen patients (72%) had macrocephaly (>2 SDS).
Eight patients (44%) had a seizure history. Six patients had febrile convulsions, whereas two had non-febrile convulsions with or without febrile convulsions. Congenital cardiac defects were found in six patients (33%); patent ductus arteriosus, ventricular septal defect, atrial septal defect and Ebstein anomaly were observed. All patients with a cardiac defect underwent a surgical correction.
Skeletal deformities were one of the most common symptoms. Thirteen (72%) patients had flat feet, ten (55.6%) patients had scoliosis and two (patients 7 and 10) had severe scoliosis (Cobb’s angle 80° and 50°, respectively) and underwent surgical correction. Chest wall deformities including pectus carinatum and excarbatum were observed in three patients. Four patients showed joint laxity.
Brain anomalies were detected by imaging studies in eight patients (44%). Four patients underwent a ventriculo−peritoneal shunt operation for hydrocephalus. The other four patients had mild ventriculomegaly and one also showed partial agenesis of the corpus callosum. Five of fourteen (35.7%) patients older than 4 years had attention deficit hyperactivity disorder and two of them had methylphenidate medication. Although three of four patients younger than 4 years showed tendency of attention deficit, they had not been diagnosed as attention deficit hyperactivity disorder because of the young age.
The height of the microdeletion group was significantly shorter than that of the intragenic mutation group (P=0.04), and age of walking alone was significantly older in the microdeletion group than that in the intragenic mutation group (P=0.02). No significant differences in the other clinical parameters were observed between the two groups, including weight/head circumference SDS, age of speaking a meaningful word, prevalence of seizures, scoliosis, cardiac anomalies, renal anomalies or flat feet.
NSD1 mutation spectrum
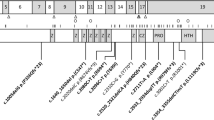
Fifteen (83%) NSD1 abnormalities were identified from the 18 unrelated patients with Sotos syndrome. The mutational spectrum is summarized in Table 2. Among the 15 patients with NSD1 abnormalities, eight (53%) had 5q35 microdeletions. The DNA sample of four patients among the eight patients with microdeletion was available for additional aCGH analysis for measuring the deletion size. All four patients had common 1.85 Mb deletion in 5q35.2q35.3, chr5:g.(175,512,657_177,360,321)del (Figure 1). The other seven (47%) had NSD1 intragenic mutations; four of them (c.4797_4798insTG, c.4591_4592insA, c.4778G>A and c.4967-1G>A) were novel mutations, and three of them (c.2227C>T, c.2362C>T and c.6349C>T) were reported previously. No familial case was observed and all mutations were de novo. Among the seven intragenic mutations, three were nonsense mutations, two were frameshift mutations, one was a missense mutation and the other was a splice site mutation. Three of the 18 patients showed neither a 5q35 microdeletion nor a NSD1 intragenic mutation.
Among the four novel mutations, the two novel nucleotide insertions (c.4797_4798insTG and c.4591_4592insA) were considered pathogenic mutations, because they lead to a frameshift and premature termination (p.Gly1600Valfs*43 and p.Met1531Asnfs*4, respectively) (* means stop codon). One novel splice site mutation (c.4967-1G>A) occurred at the end of intron 13 and could cause skipping of exon 14. The other novel missense mutation (p.Cys1593Tyr) occurred within the really interesting new gene (RING)-finger domain, which comprises 1 593 of the 2 690 amino acids of the protein. One novel variation in c.2927C>T (p.Ala976Val) was identified in patient 16.
Discussion
In this study, 15 of 18 Korean patients with Sotos syndrome had NSD1 abnormalities. This detection rate (83%) was comparable to that of previous studies performed in Japanese patients.3, 13, 17 Approximately one-half of the Korean patients had a 5q35 microdeletion, which was also similar to the results of Japanese studies.17 Although a southern Chinese study reported less frequent (12%) microdeletions, which was similar to a western study.18 It was notable that all of the four patients (patient 1, 2, 7 and 8) with microdeletion who taken aCGH showed 1.85 Mb-sized common deletion as a 1.9 Mb-sized common deletion was found in 78.7% of the patients with Sotos syndrome with the microdeletion in a previous study.16 Although the underlying reason for such a high percentage of microdeletions in Japanese and Korean patients is still unknown, the result of this study implicates very similar genetic backgrounds between Japanese and Korean. Characterization of microdeletion breakpoints in more patients of different ethnic origins may be useful to solve this issue.18 Further investigations about the breakpoint of microdeletion in a large number of Korean patients would be worth performing using a high-resolution aCGH method.
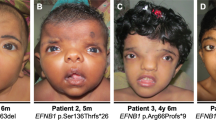
The cardinal features of Sotos syndrome were observed in almost all patients regardless of mutation type such as typical facial dysmorphism (100%), learning disabilities (100%) and overgrowth with macrocephaly (72%). The most frequently associated anomaly was skeletal deformities (72%), including scoliosis, flat feet and chest wall deformities. A total of 30% of the patients had congenital cardiac defects. Tong et al.18 reported that patients with microdeletions might be more prone to congenital heart disease but less likely to have somatic overgrowth. We also found that the height SDS was shorter (P=0.04) and age of walking alone was older (P=0.02) in the microdeletion group than those in the intragenic mutation group. However, other clinical features did not differ between the two groups in this study. Scoliosis could make it difficult to measure the exact height of patients, and the degree of scoliosis should be considered to analyze Sotos patient phenotypes. In addition, all except one intragenic mutation found in this study led to premature truncation of the NSD1 protein caused by frameshift or nonsense mutations, and severe clinical features of the intragenic mutation group might be attributed to these types of mutations. Only one patient with a NSD1 missense mutation (p.Cys1593Tyr) had less prominent facial dysmorphism and a milder learning disability (middle-school student) than those of the others. Therefore, further studies with a larger number of patients are warranted.
In terms of predicting pathogenicity of the novel missense mutation (p.Cys1593Tyr) identified in patient 1 in this study, the 1593 cysteine is a highly conserved amino acid throughout species with a unique pattern in the RING-finger domain, a specialized type of zinc-finger with 40–60 residues that bind two zinc atoms and is defined by the ‘cross-brace’ motif C-X2-C-X(9-39)-C-X(1-3)- H-X(2-3)-(N/C/H)-X2-C-X(4-48)C-X2-C (Figure 2). Many RING-finger domains simultaneously bind ubiquitination enzymes and their substrates and hence function as ligases; ubiquitination in turn targets the substrate protein for degradation.22 The novel mutation was not identified in either parents without any Sotos phenotype and was predicted to affect protein structure by the PolyPhen-2 (v2.2.2r395) program.23 Therefore, substitution of the cysteine residue by other amino acids might cause significant functional alterations in ubiquitination cascade involved in the control of gene transcription and translation.
A novel missense mutation occurred within the really interesting new gene (RING)-finger domain (p.Cys1593Tyr). The cysteine residue is a highly conserved amino acid with a unique pattern in the RING-finger domain. The figure was created using WebLogo 3 (http://weblogo.threeplusone.com/).
Patient 16 had a novel variation of c.2927C>T (p.Ala976Val), which was considered a non-pathogenic single nucleotide polymorphism because it was inherited from the unaffected mother. The PolyPhen-2 (v2.2.2r395) program also predicted it to be benign,23 though this substitution has not been reported.
Although detecting NSD1 abnormalities can confirm a diagnosis of Sotos syndrome, a negative NSD1 analysis result cannot exclude it, because the mutation detection rate is 70–80% in Japanese3, 13 and Korean patients. Thus, the clinical diagnosis is still important. In this study, three of 18 patients (17%) had no NSD1 abnormalities. Because exon deletions cannot be detected by sequencing, further testing with multiplex ligation-dependent probe amplification would be useful in these patients. Meanwhile, these patients have the possibility of having other disorders by sharing clinical features with a Sotos syndrome called Sotos-like syndrome. Yoneda et al.24 reported that microdeletions encompassing nuclear factor I-X and a nuclear factor I-X nonsense mutation have been found in individuals with clinically suspected Sotos syndrome but showed no NSD1 abnormalities. Moreover, several studies have demonstrated various genetic abnormalities including epigenetic alteration of KCNQ1T1, microduplication of 19p13.2, and a STK11 mutation in patients with Sotos-like features.25, 26, 27 Therefore, further genetic investigation is needed for the patients without NSD1 mutation, while epigenetic alteration of KCNQ1T1, which is known to be the cause of Beckwith–Wiedemann syndrome, did not be found in our three patients.
In conclusion, 15 (83%) NSD1 abnormalities were identified in 18 unrelated Korean patients with Sotos syndrome including four novel mutations. Although age of walking alone and height SDS showed significant differences in clinical features between the microdeletion and intragenic mutation groups, the small study population was the limitation of this study; thus, further studies with a larger number of patients are necessary for draw any conclusive genotype–phenotype correlations. Furthermore, additional research on genetic alterations in patients with clinically suspected Sotos syndrome with normal NSD1 is warranted.
References
Cole, T.R. & Hughes, H.E. Sotos syndrome: a study of the diagnostic criteria and natural history. J. Med. Genet. 31, 20–32 (1994).
Sotos, J.F., Dodge, P.R., Muirhead, D., Crawford, J.D. & Talbot, N.B. Cerebral gigantism in childhood - a syndrome of excessively rapid growth with acromegalic features and a nonprogressive neurologic disorder. N. Eng. J. Med. 271, 109–116 (1964).
Tatton-Brown, K., Douglas, J., Coleman, K., Baujat, G., Cole, T.R., Das, S. et al. Genotype-phenotype associations in Sotos syndrome: an analysis of 266 individuals with NSD1 aberrations. Am. J. Hum. Genet. 77, 193–204 (2005).
Saugier-Veber, P., Bonnet, C., Afenjar, A., Drouin-Garraud, V., Coubes, C., Fehrenbach, S. et al. Heterogeneity of NSD1 alterations in 116 patients with Sotos syndrome. Hum. Mut. 28, 1098–1107 (2007).
Tatton-Brown, K. & Rahman, N. Sotos syndrome. Eur. J. Hum. Genet. 15, 264–271 (2007).
Kurotaki, N., Imaizumi, K., Harada, N., Masuno, M., Kondoh, T., Nagai, T. et al. Haploinsufficiency of NSD1 causes Sotos syndrome. Nat. Genet. 30, 365–366 (2002).
Rayasam, G.V., Wendling, O., Angrand, P.O., Mark, M., Niederreither, K., Song, L. et al. NSD1 is essential for early post-implantation development and has a catalytically active SET domain. EMBO. J. 22, 3153–3163 (2003).
Aasland, R., Gibson, T.J. & Stewart, A.F. The PHD finger: implications for chromatin-mediated transcriptional regulation. Trends. Biochem. Sci. 20, 56–59 (1995).
Stec, I., Nagl, S.B., van Ommen, G.J. & den Dunnen, J.T. The PWWP domain: a potential protein-protein interaction domain in nuclear proteins influencing differentiation? FEBS Lett. 473, 1–5 (2000).
Kurotaki, N., Harada, N., Yoshiura, K., Sugano, S., Niikawa, N. & Matsumoto, N. Molecular characterization of NSD1, a human homologue of the mouse Nsd1 gene. Gene 279, 197–204 (2001).
Huang, N., vom Baur, E., Garnier, J.M., Lerouge, T., Vonesch, J.L., Lutz, Y. et al. Two distinct nuclear receptor interaction domains in NSD1, a novel SET protein that exhibits characteristics of both corepressors and coactivators. EMBO. J. 17, 3398–3412 (1998).
Cecconi, M., Forzano, F., Milani, D., Cavani, S., Baldo, C., Selicorni, A. et al. Mutation analysis of the NSD1 gene in a group of 59 patients with congenital overgrowth. Am. J. Med. Genet. A 134, 247–253 (2005).
Douglas, J., Hanks, S., Temple, I.K., Davies, S., Murray, A., Upadhyaya, M. et al. NSD1 mutations are the major cause of Sotos syndrome and occur in some cases of Weaver syndrome but are rare in other overgrowth phenotypes. Am. J. Hum. Genet. 72, 132–143 (2003).
Rio, M., Clech, L., Amiel, J., Faivre, L., Lyonnet, S., Le Merrer, M. et al. Spectrum of NSD1 mutations in Sotos and Weaver syndromes. J. Med. Genet. 40, 436–440 (2003).
Turkmen, S., Gillessen-Kaesbach, G., Meinecke, P., Albrecht, B., Neumann, L.M., Hesse, V. et al. Mutations in NSD1 are responsible for Sotos syndrome, but are not a frequent finding in other overgrowth phenotypes. Eur. J. Hum. Genet. 11, 858–865 (2003).
Visser, R., Shimokawa, O., Harada, N., Kinoshita, A., Ohta, T., Niikawa, N. et al. Identification of a 3.0-kb major recombination hotspot in patients with Sotos syndrome who carry a common 1.9-Mb microdeletion. Am. J. Hum. Genet. 76, 52–67 (2005).
Kurotaki, N., Harada, N., Shimokawa, O., Miyake, N., Kawame, H., Uetake, K. et al. Fifty microdeletions among 112 cases of Sotos syndrome: low copy repeats possibly mediate the common deletion. Hum. Mut. 22, 378–387 (2003).
Tong, T.M., Hau, E.W., Lo, I.F., Chan, D.H. & Lam, S.T. Spectrum of NSD1 gene mutations in southern Chinese patients with Sotos syndrome. Chin. Med. J. 118, 1499–1506 (2005).
Tatton-Brown, K., Cole, T.R. & Rahman, N. Sotos syndrome. in GeneReviews. http://www.ncbi.nlm.nih.gov/books/NBK1479 (University of Washington, Seattle (2012).
Nagai, T., Matsumoto, N., Kurotaki, N., Harada, N., Niikawa, N., Ogata, T. et al. Sotos syndrome and haploinsufficiency of NSD1: clinical features of intragenic mutations and submicroscopic deletions. J. Med. Genet. 40, 285–289 (2003).
Moon, J.S., Lee, S.Y., Nam, C.M., Choi, J.M., Choe, B.K., Seo, J.W. et al (2007) Korean National Growth Charts: review of developmental process and an outlook. Korean. J. Pediatr. 51, 2008.
Kevin, L.L., Jane, P.J., Shengyun, F., Albert, M.O., Shigetsugu, H. & Allan, M.W. RING fingers mediate ubiquitin-conjugating enzyme (E2)-dependent ubiquitination. Proc. Natl Acad. Sci. USA 96, 11364–11369 (1999).
Adzhubei, I.A., Schmidt, S., Peshkin, L., Ramensky, V.E., Gerasimova, A., Bork, P. et al. A method and server for predicting damaging missense mutations. Nat. Methods 7, 248–249 (2010).
Yoneda, Y., Saitsu, H., Touyama, M., Makita, Y., Miyamoto, A., Hamada, K. et al. Missense mutations in the DNA-binding/dimerization domain of NFIX cause Sotos-like features. J. Hum. Genet. 57, 207–211 (2012).
Mayo, S., Garin, I., Monfort, S., Rosello, M., Orellana, C., Oltra, S. et al. Hypomethylation of the KCNQ1OT1 imprinting center of chromosome 11 associated to Sotos-like features. J. Hum. Genet. 57, 153–156 (2012).
Lehman, A.M., du Souich, C., Chai, D., Eydoux, P., Huang, J.L., Fok, A.K. et al. 19p13.2 microduplication causes a Sotos syndrome-like phenotype and alters gene expression. Clin. Genet. 81, 56–63 (2012).
Baynam, G., Schofield, L. & Goldblatt, J. A child with an STK11 mutation and Sotos syndrome-like features: can STK11 mutations produce a Sotos syndrome phenocopy? BMJ. Case. Rep. (e-pub ahead of print 19 September 2011; doi:10.1136/bcr.07.2011.4445).
Acknowledgements
This study was supported by grant no. 2012R1A1A3001588 from the Korean Ministry of Education, Science and Technology and grant no. 04-2012-1265 from the SNUH Research Fund.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Rights and permissions
About this article
Cite this article
Bae Sohn, Y., Gon Lee, C., Min Ko, J. et al. Clinical and genetic spectrum of 18 unrelated Korean patients with Sotos syndrome: frequent 5q35 microdeletion and identification of four novel NSD1 mutations. J Hum Genet 58, 73–77 (2013). https://doi.org/10.1038/jhg.2012.135
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/jhg.2012.135
Keywords
This article is cited by
-
Drosophila NSD deletion induces developmental anomalies similar to those seen in Sotos syndrome 1 patients
Genes & Genomics (2021)
-
Identification of novel genomic imbalances in Saudi patients with congenital heart disease
Molecular Cytogenetics (2018)