Abstract
Ectodermal-dysplasia-syndactyly syndrome (EDSS1) is a rare form of ectodermal dysplasia (ED), affecting skin and its appendages mainly hair, teeth and nails. In the present study, we have investigated a large consanguineous Pakistani family with 10 individuals showing features of EDSS1. Human genome was screened using highly polymorphic microsatellite markers to identify the gene causing EDSS1. The disease locus for EDSS1 was assigned to chromosome 1q23.1-q23.3. This region corresponds to 5.63 Mb according to the sequenced based physical map (Build 36.2) of the human genome and flanked by markers D1S1653 and D1S1677. A maximum two-point LOD score of 5.05 was obtained with the marker D1S484. Sequence analysis revealed a homozygous missense mutation (c.635C>G; p.Pro212Arg) in the recently reported PVRL4 gene causing EDSS1. The involvement of mutant nectin-4 in causing EDSS1 may open up interesting prospectives into the role of cell adhesion molecules in causing syndromic forms of EDs.
Similar content being viewed by others
Introduction
Ectodermal dysplasias (EDs) are a group of congenital disorders that share common abnormalities of at least two or more ectodermal derivatives such as hair, teeth, nails and sweat glands. To date, more than 200 conditions have been described under the term ED. EDs may be of pure forms involving only classical ectodermal appendages or syndromic forms affecting other organs of the body as well. Syndromic forms of EDs are mostly associated with deafness, skeletal defects, mental retardation, ichthyosis, palmoplantar keratoderma, eye abnormalities, facial deformities, cleft lip and palate, and other systemic conditions.
Ectodermal-dysplasia-syndactyly syndrome (EDSS1, MIM 613573) is a rare form of ED affecting skin and its appendages mainly hair, teeth and nails. Tariq et al.1 have described a large Pakistani family with four individuals affected with ectodermal-dysplasia-cutaneous syndactyly syndrome (EDSS2, MIM 613576). All these individuals showed features of hypotrichosis, hypoplastic nails, tooth enamel hypoplasia, hyperhidrosis, palmoplantar keratoderma and bilateral partial cutaneous syndactyly. Further these authors mapped a causative gene for this syndrome on chromosome 7p21.1-p14.3. Some of the clinical features observed in the family reported by Tariq et al.1 were also reported in patients of two most clinically related conditions, EDSS12 and Baisch's3 syndrome. Very recently, Brancati et al.4 described an Algerian and an Italian family showing features similar to those reported by Tariq et al.1 Brancati et al.4 named the ED syndrome observed in the two families as ectodermal dysplasia syndactyly syndrome (EDSS1). These authors identified nectin-4 encoding PVRL4 gene on chromosome 1q23.1-q23.3 causing EDSS1 phenotype.
The present study described a large consanguineous Pakistani family with EDSS1 phenotypes, originated from a remote area of Punjab province Pakistan. By homozygosity mapping we assigned the disease locus on chromosome 1q23.1-q23.3. DNA sequence analysis detected a homozygous missense mutation in PVRL4 gene.
Materials and methods
Human subjects
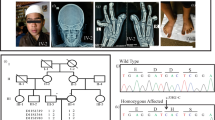
The present five-generation family, exhibiting features of EDSS1 (Figure 1), belongs to a remote region in Punjab province of Pakistan. The affected individuals were born of first cousin parents with variable degree of consanguinity (parents IV-1, IV-2 and IV-5, IV-6 and IV-4, IV-7 were first cousins whereas III-5, IV-3 were first cousins one removed), suggesting that affected individuals are homozygous for a mutant allele. All affected and unaffected individuals underwent examination at the local Government Hospital. Before the start of the study, approval was obtained from Institutional Review Board (IRB), Quaid-i-Azam University, Islamabad, Pakistan. Informed consent for the study including presentation of the photographs for publication was obtained from affected individuals and their parents.
Haplotype analysis of the family segregating autosomal recessive ectodermal dysplasia-syndactyly syndrome (EDSS1). Affected males and females are indicated by filled squares and circles, respectively. Symbols with crossed lines indicate the deceased individuals. Double lines between individuals represent consanguineous unions. For each individual, haplotypes of the most closely linked microsatellite markers are shown below the symbol. The genetic positions (centiMorgan) and arrangement of microsatellite markers is according to the Rutgers combined linkage physical map build 36.2.8
Extraction of genomic DNA and genotyping
Venous blood samples were collected from nine affected (V-2, V-3, V-4, V-6, V-7, V-8, V-10, V-14, V16) and 11 unaffected (III-5, IV-1, IV-2, IV-4, IV-5, IV-6, IV-7, V-1, V-12, V-13, V-15) members of the family in EDTA containing vacutainer sets.
Genomic DNA from peripheral white blood cells was extracted using a commercially available DNA purification kit (MBI Fermentas, Life Sciences, York, UK). The DNA was quantified by Nanodrop-1000 spectrophotometer (Thermo Scientific, Wilmington, NC, USA) readings at optical density of 260 nm and diluted to 40 ng μl−1 for PCR amplification. The PCR reaction was performed in a total volume of 25 μl containing 40 ng of human genomic DNA, 10-20 pmol of each primer, one unit of Taq DNA polymerase, 0.2 mM of dNTP mix, 1 mM MgCl2 and 2.5 μl of 10 × NH4SO4 buffer (MBI Fermentas, Life Sciences). For DNA amplification, standard temperature conditions of PCR were followed.
Homozygosity mapping
Initially, all the affected and unaffected individuals of the family were genotyped using 5–6 highly polymorphic microsatellite markers linked to several previously reported ED phenotypes. These included EDSS2 on chromosome 7p21.1-p14.3 (D7S488, D7S2562, D7S2190, D7S1808, D7S2491, D7S817), excision-repair complimenting defective in Chinese hamster 3 (ERCC3, MIM 133510) gene on chromosome 2q21 (D2S1328, D2S2339, D2S2271, D2S2215, D2S1260), wingless-type MMTV integration site family member 10A (WNT10A, MIM 606268) gene on chromosome 2q35 (D2S2248, D2S1338, D2S2244, D2S163, D2S126), fibroblast growth factor receptor 3 (FGFR3, MIM 134934) gene on chromosome 4p16.3 (D4S111, D4S3038, D4S115, D4S1614, D4S126), fibroblast growth factor 10 (FGF10, MIM 602115) gene on chromosome 5p13-p12 (D5S430, D5S665, D5S1396, D5S2106, D5S1388), general transcription factor 2H, polypeptide 5 (GTF2H5, MIM 608780) gene on chromosome 6q25.3 (D6S947, D6S442, D6S363, D6S969, D6S1581), trichorhinophalangeal syndrome 1 (TRPS1, MIM 190350) gene on chromosome 8q24.12 (D8S565, D8S1694, D8S384, D8S199, D8S1101), fibroblast growth factor receptor 2 (FGFR2, MIM 176943) gene on chromosome 10q26 (D10S542, D10S1722, D10S1483, D10S587, D10S2322), poliovirus receptor-like 1 (PVRL1, MIM 600644) gene on chromosome 11q23 (D11S4104, D11S924, D11S994, D11S925, D11S4089), type II keratin genes cluster on chromosome 12q13.13 (D12S270, D12S1604, D12S103, D12S1724, D12S1632), forkhead box N1 (FOXN1 or WHN, MIM 600838) gene on chromosome 17q11-q12 (D17S841, D17S1294, D17S1800, D17S798, D17S1850), distal-less homoebox 3 (DLX3, MIM 600525) gene on chromosome 17q21.3-q22 (D17S943, D17S747, D17S1865, D17S752, D17S1607) and excision-repair complimenting defective in Chinese hamster 2 (ERCC2, MIM 126340) gene on chromosome 19q13.2-q13.3 (D19S190, D19S554, D19S223, D19S537, D19S913). Examination of the haplotypes generated by SIMWALK25 did not reveal any region of homozygosity among affected individuals of the family.
After exclusion of the family from linkage to the known genes, the family was subjected to human genome scan (22 autosomes and sex chromosomes) to map a causative gene. The genome-wide scan was conducted on DNA samples from three affected (V-2, V-6, V-10) and one unaffected (IV-1) individual. A total of 534 short tandem repeat polymorphic microsatellite markers, with an average heterozygosity of 0.75 from Linkage Mapping Set (Invitrogen, San Diego, CA, USA), were genotyped in the present genome scan. These markers are spaced ∼6–7 cM apart on 22 autosomes, and X and Y chromosomes. The amplified PCR products were resolved on 8% non-denaturing polyacrylamide gel and visualized by ethidium bromide staining to score the alleles by manual inspection. Allele size of microsatellite markers was determined using 10, 20 and 50-bp DNA ladders (MBI Fermentas, Life Sciences).
Statistical analysis
PEDSTATS6 was used to evaluate the genotyped data for Mendelian incompatibilities and to assess the data for occurrence of unlikely genotypes. SIMWALK25 was used to construct the haplotypes. Parametric linkage analysis was carried out using the online version of superlink software (http://bioinfo.cs.technion.ac.il/superlink-online/) and MLINK program of FASTLINK7 computer package. Two-point LOD scores were calculated using a fully penetrant autosomal recessive model with an assumed disease allele frequency of 0.0001. The LOD score values, shown in Table 1, were obtained with the assumption that the allele frequencies of the studied polymorphic markers were equal because it was not possible to estimate allele frequencies from the founders, as these markers were only genotyped in this family. The National Center for Biotechnology Information (NCBI) build 36.2 sequence-based physical map8 was used to determine the order of the genome scan and fine mapping markers.
Mutational analysis
Entire coding region and splice junction sites of PVRL4 gene were amplified by PCR and screened by DNA sequencing for potential sequence variants. Primer sequences were designed for each exon using Primer3 software9 and checked for specificity using Basic Local Alignment Search Tool (BLAST; http://www.ncbi.nlm.nih.gov/blast). Purification of the PCR-amplified DNA was performed with commercially available kits (Marligen Biosciences, Rockville, MD, USA). Sequencing of the PVRL4 gene was performed following dideoxy chain termination method and using Big Dye Terminator v3.1 Cycle Sequencing Kit, together with an ABI Prism 310 Genetic Analyzer (Applera, Foster City, CA, USA). Sequence variants were identified via Bioedit sequence alignment editor version 6.0.7 (www.mbio.ncsu.edu/BioEdit/bioedit.html).
Results
Clinical findings
The family, presented here, has 10 individuals affected with EDSS1. All the affected individuals showed features of sparse scalp hair, sparse to absent eyebrows and eyelashes (Figure 2a). Conical and cylindrical shaped teeth with ill-defined surface morphology and enamel hypoplasia were observed in both arches of the affected individuals (Figure 2b). Hypoplastic finger- and toe-nails with nail-plate degeneration of variable severity and hyperkeratosis over the palms was noted in most of the affected individuals (Figures 2c and d). All affected individuals manifested bilateral inter-digital webbing (cutaneous syndactyly) of third and fourth fingers in the hand, and second and third toe digits (Figures 2d and e). Radiological examinations (not shown) revealed that fusion of the digits is restricted to skin and has not affected the bones.
Clinical features of EDSS1. Sparse scalp hair, sparse eyebrows and eyelashes, and normal moustache and beard hair in patient V-10 (a). Conical/cylindrical malpositioned teeth in patient V-14 at 15 years of age (b). Hypoplastic nails showing degenerate nail-palate in patient V-2 (c). Cutaneous syndactyly of the third and fourth digits of the hands in patient V-8 (d) and of the second and third digits of the feet in patient V-14 (e).
Abnormalities of heart and other systems including respiratory, urinogenital and endocrinal were not observed in any of the affected individual. They were mentally normal facing no speech problem. Musculoskeletal system appeared normal in radiological examinations. Ophthalmological examinations revealed normal functioning of the eyes. No evidence of hypo- or hyper-hidrosis was observed on any part of the body of the affected individuals.
Mapping ectodermal-dysplasia-cutaneous syndactyly syndrome on chromosome 1q23.1-q23.3
Initially, linkage of the present family was tested to several genes reported earlier to cause related phenotypes. These included EDSS2 on chromosome 7p21.1-p14.3, ERCC3 on chromosome 2q21, WNT10A on chromosome 2q35, FGFR3 on chromosome 4p16.3, FGF10 on chromosome 5p13-p12, GTF2H5 on chromosome 6q25.3, TRPS1 on chromosome 8q24.12, FGFR2 on chromosome 10q26, PVRL1 on chromosome 11q23, type II keratin genes cluster on chromosome 12q13.13, FOXN1 on chromosome 17q11-q12, DLX3 on chromosome 17q21.3-q22 and ERCC2 on chromosome 19q13.2-q13.3.
After exclusion of the family from linkage to the known genes, the family was subjected to human genome scan (22 autosomes and sex chromosomes) to map a causative gene. The genome scan was conducted on DNA samples from three affected (V-2, V-6, V-10) and one unaffected (IV-1) individual. A total of 530 microsatellite markers were genotyped in the genome scan. The affected individuals showed homozygosity with five microsatellite markers including D1S398 (1q23.2), D3S3566 (3p14), D9S1867 (9q21), D16S415 (16p12) and D17S809 (17q21). Upon testing DNA from other 16 members including six affected (V-3, V-4, V-7, V-8, V-14, V-16) and ten unaffected (III-5, IV-2, IV-4, IV-5, IV-6, IV-7, V-1, V-12, V-13, V-15) of the family, linkage to four of these regions was excluded. All affected individuals of the family were found to be homozygous at marker D1S398 located at 161.98 cM on chromosome 1q23.2. Unaffected individuals of the family were heterozygous at this marker.
In order to fine map the region, 14 additional microsatellite markers (D1S2624, D1S1653, D1S2635, D1S1167, D1S2707, D1S2771, D1S484, D1S2705, D1S1679, D1S2844, D1S1677, D1S104, D1S403, ATA38A05) were selected and genotyped in all affected and unaffected individuals of the family. Four markers (D1S2635, D1S1167, D1S2707, D1S2771) were non-informative in the family and therefore not considered for further analysis. Analysis of the fine mapping markers genotypes within this region by PEDSTATS6 did not elucidate any genotyping errors. The results of the two-point linkage analysis using MLINK program of FASTLINK7 computer package and the online version of superlink software (http://bioinfo.cs.technion.ac.il/superlink-online/) are presented in Table 1. The highest two-point LOD score, at zero recombination fraction (θ=0.00), of 5.05 was obtained with marker D1S484.
Haplotypes using SIMWALK25 were constructed to determine the critical recombination events in the family (Figure 1). A recombination event between markers D1S1653 and D1S398 occurred in two-affected individual (V-2, V-3) defined the centromeric boundary of the linkage interval. The telomeric boundary of the interval was defined by recombination events occurred between marker D1S2844 and D1S1677 in two affected individuals (V-10, V-14). The linkage interval flanked by markers D1S1653 (160.09 cM) and D1S1677 (170.25 cM) contains 5.63 Mb physical distance, which is 10.16 cM long according to the Rutgers combined linkage-physical map build 36.2 of the human genome.8
Screening PVRL4 gene
At this time when a novel locus for EDSS1 was mapped in the present family, Brancati et al.4 identified a poliovirus receptor-related 4 (PVRL4, MIM 609607) gene, on chromosome 1q23.1-q23.3, causing EDSS1 in Algerian and Italian families. The linkage interval of 5.63 Mb of EDSS1 mapped on chromosome 1q23.1-q23.3, in the present family, contains the same PVRL4 gene. Therefore, the PVRL4 gene, encoding adhesion receptor nectin-4 protein, was selected as the most plausible candidate in the present family.
To search for potential sequence variants, coding exons and splice junction sites of PVRL4 gene were sequenced initially in two affected individuals and one unaffected individual of the family. A homozygous C to G transition at complementary DNA position 635 (c.635C>G) was detected in exon 3 of PVRL4 gene in both the affected individuals (Figure 3). This transition exchanged a proline to arginine residue at amino-acid position 212 (p.Pro212Arg) of the PVRL4 protein. Sequencing of exon 3 of the gene in all the rest of the members of the family revealed the presence of the mutation in homozygous state only in affected individuals. The genetic change (c.635C>G; p.Pro212Arg) was present in a heterozygous state in the obligate heterozygous carriers of the family. To ensure that the mutation does not represent a neutral polymorphism in this population, a panel of 200 unrelated, unaffected, ethnically matched control individuals was screened for the mutation, and it was not identified outside the family.
Discussion
The present study described a large consanguineous family originated from a remote region in Punjab province of Pakistan. All nine affected individuals of the family shared clinical features compatible with EDSS1. The clinical features observed in affected individuals in the present family overlap with the affected individuals of the families reported earlier.1, 4 The features such as hypotrichosis, teeth anomalies and cutaneous syndactyly of hands and feet, observed in affected individuals of the present family, are similar to those reported in another Pakistani family with EDSS2 by Tariq et al.,1 and very recently in Algerian and Italian families with EDSS1 by Brancati et al.4 However, the hair morphology representing repeated twists of the hair (pili torti) reported in the family with EDSS14 were not observed in the present and the family reported with EDSS2.1 Other clinical features including mild keratoderma and nail dystrophy, found in the present family, were also reported by Tariq et al.1 in the Pakistani family but not by Brancati et al.4 in Algerian and Italian families.
In the present family, human genome scan mapped the EDSS1 to chromosome 1q23.1-1q23.3. This locus is different from the one mapped earlier on chromosome 7p21.1-p14.3 by Tariq et al.1 The novel EDSS1 locus on chromosome 1q23.1-1q23.3 spans the genetic interval of 10.16 cM corresponding to 5.63 Mb according to the Rutgers combined linkage-physical map build 36.2 of the human genome.8 This region contains several genes. While selection of genes for screening was underway, Brancati et al.4 reported the involvement of a poliovirus receptor-related 4 (PVRL4) gene, on chromosome 1q23.1-1q23.3, in causing EDSS1. These authors identified mutations in PVRL4 gene in two families that belonged to Algeria and Italy. In an Algerian family a missense mutation (p.Arg284Gln) and in an Italian family a compound heterozygous mutation (p.Thr185Met, p.Pro304HisfsX2) was identified. Therefore, the same PVRL4 gene was sequenced in the affected and unaffected individuals of the family investigated here. DNA sequence analysis detected a novel homozygous missense mutation (c.635C>G, p.Pro212Arg) in PVRL4 gene in affected individuals of the family.
The poliovirus receptor-related 4 (PVRL4) gene contains nine exons encoding adhesion receptor nectin-4, which is a 55.5 kDa protein containing 510 amino acids.10 According to protein knowledge database UniProtKb (http://www.uniprot.org/uniprot/Q96NY8), nectin-4 contains an N-terminal signal peptide (1–31 amino acids), an extracellular domain (32–349 amino acids) having three immunoglobulin-like sub-domains (V-type1 32–144 amino acids, C2-type1 148–237 amino acids, C2-type2 248–331 amino acids), a transmembrane domain (350–370 amino acids) and a cytoplasmic domain (371–510 amino acids). The signal peptide is encoded by exon 1, V-type1 domain by exon 2, C2-type1 domain by exons 3, C2-type2 domain by exon 4 and 5, transmembrane domain by exon 6 and cytoplasmic domain by exon 7, 8 and 9. The missense mutation (p.Pro212Arg) identified in the present family resides in the second immunoglobulin-like (C2-type1) domain of nectin-4 protein. Multiple Sequence Alignment via ClustalW (http://www.ebi.ac.uk/Tools/clustalw2) of nectin-4 protein from different vertebrate species revealed that proline at amino-acid position 212 is a conserved residue (Figure 4). PolyPhen (polymorphism phenotyping) software (http://genetics.bwh.harvard.edu/pph/) predicts that replacement of proline (helix breaker) by arginine residue disrupt the structure of the protein. PSIC (Position Specific Independent Counts) score difference for normal and mutant allele in PVRL4 gene in the present case is 2.325. The PolyPhen is a computational tool for the prediction of functional mutations and computes the absolute value of difference between profile scores of both allelic variants in the polymorphic position. Big values of this difference may indicate that the studied substitution is never observed in the protein.
(a) Schematic view of the nectin-4 protein exhibiting three extracellular immunoglobulin (Ig)-like domains, a transmembrane domain and cytoplasmic domain. The mutation (p.Pro212Arg) resides in the second Ig-like domain (C2-type1) of nectin-4 protein. S and TM represent signal peptide and transmembrane domain, respectively. The numbers below represent the amino acids located at the boundaries of each domain. (b) Amino-acid sequence alignment showing conservation of Pro212 among different species.
Epithelial cell–cell contacts are mainly dependent on three major adhesive structures namely tight junctions, adherens junctions and desmosomes. Several adhesion proteins of these three types of junctions connect the neighboring cells through homophilic and heterophilic adhesive interactions. Two major classes of adhesive protein complexes cadherin-catenin and nectin-afadin of adherens junction are involved in regulating tissue formation and morphogenesis during development and maintenance of solid tissues in the adult organism.11
Nectins are immunoglobulin-like family of proteins containing four members. Polio virus-like receptor (PVRL4) encodes nectin-4, a cell adhesion molecule mainly involved in the arrangement of cadherin-based adherens junctions. Nectin-4 shows significant expression in skin, hair follicles and cultured keratinocytes.10 Brancati et al.4 determined the critical role of nectin-4 in hair morphogenesis and cycling. These authors’ have further shown presence of nectin-4 at cell–cell junctions of human keratinocytes, spinous to the granular layer of epidermis and in all the non-keratinized structures of hair with a stronger signal within inner root sheath layers and hair shaft cortex. In addition, Brancati et al.4 showed inter-digital expression of nectin-4 at embryonic stage E15.5 in mouse.
Nectin-4 is the second member of the nectin family of proteins involved in human genetic disorders. Previously, mutations in nectin-1 encoded by PVRL1 gene have been shown to cause cleft lip-palate ED (CLPED1, MIM 225069).12 As described by Brancati et al.4 similarity in the expression pattern of nection-1 and nectin-4 and the trans-heterophilic interaction between these two proteins predicts a common mode of action of these molecules in regulating ectodermal organogenesis.10 Brancati et al.4 have further shown that mutant nectin-4 affects both nectin-afadin and cadherin-catenin complexes, which leads to defective organogenesis.
References
Tariq, M., Khan, M. N. & Ahmad, W. Ectodermal dysplasia-cutaneous syndactyly syndrome maps to chromosome 7p21.1-p14.3. Hum. Genet. 125, 421–429 (2009).
Boudghene-Stambouli, O. & Merad-Boudia, A. Association of ectodermal dysplasia and syndactylia. Ann. Dermatol. Venereol. 118, 107–110 (1991).
Freire-Maia, N. & Pinheiro, M. Ectodermal dysplasias: a clinical and genetic study. Alan. R. Liss. New York 172–173 (1984).
Brancati, F., Fortugno, P., Bottillo, I., Lopez, M., Josselin, E., Boudghene-Stambouli, O. et al. Mutations in PVRL4, encoding cell adhesion molecule nectin-4, cause ectodermal dysplasia-syndactyly syndrome. Am. J. Hum. Genet. 87, 265–273 (2010).
Sobel, E. & Lange, K. Descent graphs in pedigree analysis: applications to haplotyping, location scores, and marker-sharing statistics. Am. J. Hum. Genet. 58, 1323–1337 (1996).
Wigginton, J. E. & Abecasis, G. R. PEDSTATS: descriptive statistics, graphics and quality assessment for gene mapping data. Bioinformatics 21, 3445–3447 (2005).
Cottingham##Jr, R. W., Idury, R. M. & Schaffer, A. A. Faster sequential genetic linkage computations. Am. J. Hum. Genet. 53, 252–263 (1993).
Matise, T. C., Chen, F., Chen, W., De-La-Vega, F. M., Hansen, M., He, C. et al. A second-generation combined linkage physical map of the human genome. Genome Res. 17, 1783–1786 (2007).
Rozen, S. & Skaletsky, H. Primer3 on the WWW for general users and for biologist programmers. Methods Mol. Biol. 132, 365–386 (2000).
Reymond, N., Fabre, S., Lecocq, E., Adelaide, J., Dubreuil, P. & Lopez, M. Nectin4/PRR4, a new Afadin-associated member of the nectin family that trans-interacts with nectin1/PRR1 through V domain interaction. J. Biol. Chem. 276, 43205–43215 (2001).
Perez-Moreno, M., Jamora, C. & Fuchs, E. Sticky business: orchestrating cellular signals at adherens junctions. Cell 112, 535–548 (2003).
Suzuki, K., Hu, D., Bustos, T., Zlotogora, J., Richieri-Costa, A., Helms, J. A. et al. Mutations of PVRL1, encoding a cell-cell adhesion molecule/herpesvirus receptor, in cleft lip/palate-ectodermal dysplasia. Nat. Genet. 25, 427–430 (2000).
Acknowledgements
We are grateful to all the members of the family for their invaluable participation and cooperation. Funding: the work presented here was funded by Higher Education Commission (HEC), Islamabad, Pakistan. Musharraf Jelani was supported by an indigenous PhD fellowship from HEC, Islamabad, Pakistan.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Rights and permissions
About this article
Cite this article
Jelani, M., Chishti, M. & Ahmad, W. Mutation in PVRL4 gene encoding nectin-4 underlies ectodermal-dysplasia-syndactyly syndrome (EDSS1). J Hum Genet 56, 352–357 (2011). https://doi.org/10.1038/jhg.2011.18
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/jhg.2011.18
Keywords
This article is cited by
-
The biology and rationale of targeting nectin-4 in urothelial carcinoma
Nature Reviews Urology (2021)
-
Chemovirotherapeutic Treatment Using Camptothecin Enhances Oncolytic Measles Virus-Mediated Killing of Breast Cancer Cells
Scientific Reports (2019)
-
Potential role for nectin-4 in the pathogenesis of pre-eclampsia: a molecular genetic study
BMC Medical Genetics (2018)
-
Epidermal cell junctions and their regulation by p63 in health and disease
Cell and Tissue Research (2015)
-
Nectin-4 Mutations Causing Ectodermal Dysplasia with Syndactyly Perturb the Rac1 Pathway and the Kinetics of Adherens Junction Formation
Journal of Investigative Dermatology (2014)