Abstract
Reflux esophagitis (RO) and Barrett's esophagus (BO) can cause esophageal adenocarcinoma (OAC). The esophageal mucosa in the RO–BO–OAC cascade is chronically exposed to gastro-esophageal reflux. Epidermal growth factor (EGF) has an important role in the protection and repair of mucosal damage, and non-physiologic levels are associated with gastrointestinal tumors. The aim is to determine the functional effect of EGF gene polymorphisms on RO, BO and OAC development. A cohort of 871 unrelated Dutch Caucasians consisted of 198 healthy controls, 298 RO patients, 246 BO patients and 129 OAC patients. The frequency of the EGF-production-associated 5′UTR A+61G polymorphism was determined in these four groups. EGF immunohistochemistry was performed on BO biopsies. EGF expression was significantly lower in the G/G genotype compared with the A/G (P=0.008) and A/A (P=0.002) group. The G/G genotype was significantly more prevalent in RO (odds ratios (OR)=2.6; 95% confidence intervals (95% CI): 1.3–5.2), BO (OR=3.0; 95% CI: 1.5–6.2) and OAC (OR=4.1; 95% CI: 1.8–9.7) than in controls. The G allele is associated with reduced EGF expression and increased risk for RO, BO and OAC development. This indicates that reduced mucosal protection resulting from genetically decreased EGF expression enhances esophageal tumor development.
Similar content being viewed by others
Introduction
Esophageal adenocarcinoma (OAC) is rising faster than any other epithelial malignancy in the Western world since 1970.1 OAC frequently arises from reflux esophagitis (RO) and Barrett's esophagus (BO), a chronic inflammatory condition characterized by a change in the normal esophageal epithelium into intestinal metaplasia as a result of persistent severe reflux,2, 3, 4 and genomic instability.5
Several twin studies have provided evidence for a genetic component in RO.6, 7 The presence of an inherited genetic component impacting on the individual predisposition to develop BO has been accumulated over the last three decades,8 and the only predisposing factor in families with BO and OAC seems to be RO.9, 10
The stratified squamous epithelium of the healthy esophagus possesses a variety of intrinsic defenses that enable it to resist acid-peptic reflux, divided in pre-epithelial defense, epithelial defense and post-epithelial defense.11, 12 Growth factors such as epidermal growth factor (EGF) and transforming growth factor-α are associated with epithelial proliferation and restitution that are the two key mechanisms involved in esophageal epithelial defense against acid, maintaining epithelial integrity and enabling rapid repair after injury.13 Multiple signaling pathways are activated by binding of transforming growth factor (TGF)-α and EGF with their receptor EGFR,14, 15 resulting in proliferation and differentiation of epithelial tissues.16, 17
Basal EGF levels are in part determined at genetic level, and variation at a single-nucleotide polymorphism (SNP) within the EGF gene has been associated with altered EGF production.18 This SNP in the 5′ UTR of the EGF gene has been associated with the risk of various tumors, such as OAC, gallbladder cancer, ovarian cancer and gastric cancer.19, 20, 21, 22 For all three malignancies, it was reported that hetero- and homozygous G allele carriers had an increased risk for cancer compared with non-carriers. However, these associations were in contrary to others that showed an increased risk for A allele carriers with renal cell carcinoma, gastric cancer and esophageal squamous cell carcinoma.18, 21, 23
Several studies have reported on the expression of TGF-α, EGF and EGFR in the human esophageal mucosa, both at the level of RNA and at protein level.24, 25, 26, 27 EGF expression in BO was significantly higher compared with both squamous epithelium and gastric mucosa, while EGF expression was depleted in RO.24 BO showed local EGF expression and an overexpression of both TGF-α and EGFR suggesting a deregulation of important proliferation control mechanisms in these epithelial cells. EGFR overexpression in OAC correlated with advanced pathologic tumor classification and lymph node metastasis.28
BO is an ideal model to study cancer genetics, and EGF is suggested to be involved in esophageal cancer development.29 We postulated that the functional EGF 5′ UTR G allele is associated with individual susceptibility for RO, BO or OAC, and affects local EGF expression in the esophagus.
Materials and methods
Study design
Between November 2002 and March 2006, all subsequent patients with RO, BO and OAC who visited the endoscopy unit of the Erasmus MC—University Medical Centre Rotterdam or the IJsselland Hospital, Capelle aan den IJssel were invited to participate in this study. The overall response rate was 85%. Participants underwent upper endoscopy for evaluation of reflux-related symptoms, surveillance of BO, or odynophagia or dysphagia. The presence of RO was confirmed at endoscopy, and the length of any columnar-lined segment was determined by measuring the distance between the squamo-columnar junction and the proximal margin of the longitudinal gastric folds. Participants were only included if they had (i) RO without the presence of BO, (ii) BO defined as a columnar-lined segment in the esophagus of ⩾2 cm in length with specialized intestinal metaplasia at histology found in at least one of the biopsies or (iii) OAC defined at histology as an adenocarcinoma in BO or at the gastro-esophageal junction with >50% of the tumor mass in the esophagus. The response rate of the patients was 86.3%. Not all biopsies fitted the above-mentioned histological criteria, therefore ∼50% of RO patients could be included, 90% of BO patients and 95% of OAC patients.
Between October 2004 and April 2005, healthy volunteers (referred to as normal controls) were recruited from the general population of general practitioners practices. Subsequent adult patients visiting the participating general practices were examined by means of a validated questionnaire. Subjects were eligible for inclusion if they never had reflux complaints, retrosternal pain, or regurgitation, nor used antacids, H2-receptor antagonists or proton pump inhibitors. We performed age, sex and race frequency matching during the selection of controls. The response rate was 48.9%.
All individuals of the four study groups donated 10 ml of whole blood.
The study was approved by the local institutional review boards of both participating hospitals. All participants were genetically unrelated Dutch Caucasian people, and aged over 18. Before inclusion, all participants signed a written informed consent.
Genotyping of the EGF 5′ UTR polymorphism
Genomic DNA was extracted from 5 ml of whole blood by standard procedures (Wizard Genomic DNA Purification kit; Promega, Madison, WI, USA). SNP analysis of EGF (+61A/G) genotypes was performed by a technician unaware of the clinical findings for these patients on 20 ng DNA (Kbiosciences, Herts, UK) with a competitive allele-specific PCR system30 using primers designed in the flanking region of SNP CAAGGGTTGT[A/G]GCTGGAACTTTCCATCAGT located at position +61 in the 5′ UTR (rs4444903).
Immunohistochemical analysis of EGF
The local institutional review board only allowed us to use remnants of routine biopsy samples from the Erasmus MC—University Medical Centre Rotterdam. From the 246 BO patients who participated in this study, only 37 BO patients had sufficient high-quality biopsy material of the BO segment left to allow an immunohistochemical study of esophageal EGF and EGFR expression. Paraffin samples were cut at 4 μm, deparaffinized in xylene, hydrated using a graded series of alcohol, and stained with hematoxylin and eosin. The two sequential paraffin slides directly following the slide used for histological evaluation were used for EGF and EGFR analysis. The local institutional review board did not allow extensive testing for the presence of EGF and EGFR levels in OAC biopsy samples, because a preliminary screening (see results) of four samples revealed that overexpression of both EGF and EGFR was to be expected in all OAC samples. For EGF staining, endogenous peroxidase activity was inactivated with 0.3% H2O2 in 100% methanol for 30 min. Subsequently, antigen retrieval was performed by boiling the slides in 10 mM monocitric acid (pH 6.0) for 15 min, and allowing them to slowly cool down to room temperature. Antigen retrieval for EGFR was performed by 5 min incubation with Prot K at room temperature, followed by one washing with ice-cold sterile PBS. The slides were then blocked with 10% normal human plasma for 20 min at room temperature. The sections were stained using a primary antibody against EGF (anti-EGF clone EGF-10; 1:200 dilution; Sigma, St Louis, MO, USA) or EGFR (anti-EGFR clone F4; 1:250 dilution; Sigma). Binding of the primary antibody was visualized by the addition of anti-mouse Envision labeled with horseradish peroxidase (DAKO, Glostrup, Denmark).
Two independent observers (VM and KvZ) evaluated the sections of the EGF and EGFR stainings, while blinded for the score of the other observer, and unaware of the patient characteristics and genotype. To determine EGF and EGFR expression, the number of stained cells was scored per microscopic field as 0 (negative), 1 (mild, 1–100 positive cells per field) and 2 (strong, >100 positive cells per field). The slides were evaluated using a Zeiss microscope (Axioskop 20, Carl Zeiss, Sliedrecht, The Netherlands) with a standard magnification (200 × ), and the images were recorded with a Nikon camera (DS-5M-U1) and Nikon Eclipse Net 2000 software (Nikon, Badhoevedorp, The Netherlands). The mean variation in scores between observers was found to be <5%, and the average of the two observations was used to analyze the relation with the EGF genotypes.
Statistical analysis
The study was powered (80%) to allow detection of a 10% difference in allele distribution between the patient groups (significance level 5%). Differences between allele distributions of the EGF 5′ UTR polymorphism, as well as differences between the patient groups in number, age and sex were determined by χ2 analysis. Age and sex-corrected odds ratios (OR) and 95% confidence intervals (95% CI) were calculated for the association between healthy controls and RO, BO or OAC, respectively, by logistic regression analysis. Disease causality was calculated with ordinal regression for the RO–BO–OAC cascade in relation to sex. The mean immunohistochemical EGF expression was compared between the three genotypes with analysis of variance. A difference between the median immunohistochemical EGF expression per genotype was calculated with the Wilcoxon non-ranked sum test. A two-sided P-value ⩽0.05 was considered to be statistically significant. All statistical analyses were conducted with the SPSS software package v11.1 (SPSS, Chicago, IL, USA).
Results
Patient characteristics
In total, 871 participants were included; 198 controls, 298 RO, 246 BO and 129 OAC patients. Table 1 shows that male gender was more common in BO and OAC than among controls and RO patients. On average, patients with BO and OAC were older than controls and RO patients. When comparing the OAC with the BO group, slightly higher age and higher prevalence of males were found in the OAC patients.
G/G genotype is more prevalent among RO, BO and OAC patients
Using controls as a reference, the association of an EGF polymorphism was tested for RO, BO or OAC patients. Allele frequencies for the EGF 5′ UTR polymorphism among the 198 controls were 32.8% (G) and 67.2% (A), respectively. The allele frequency of the major allele (A) was 60.9% in RO, 59.3% in BO and 58.1% in OAC.
The distribution of genotype frequencies for the polymorphism investigated was consistent with Hardy–Weinberg equilibrium in both the patient and control groups (P>0.05). Comparing the patient groups with the controls showed that EGF G/G homozygosity was observed more frequently in RO (OR=2.62; 95% CI: 1.33–5.15), BO (OR=2.85; 95% CI: 1.42–5.75) and OAC (OR=3.12; 95% CI: 1.42–6.85) compared with controls. The adjustment for age and sex showed that EGF 5′ UTR G/G homozygosity was significantly more common in RO (OR=2.61; 95% CI: 1.32–5.15), BO (OR=3.04; 95% CI: 1.49–6.17) and OAC (OR=4.12; 95% CI: 1.76–9.65) compared with the controls (Figure 1). There was no significant difference in the frequency distribution of EGF G/G genotype in BO versus RO (OR=1.00; P=0.74), in OAC versus RO (OR=1.46; P=0.35) and OAC versus BO (OR=1.19; P=0.65). Figure 1 shows that the frequency of A/A genotype decreases gradually for RO, BO and finally for OAC, while there is a clear increase for the G/G genotype frequency. This G allele shift taken together with the odds ratios for RO (P=0.006), BO (P=0.002) and OAC (P=0.001) indicates that the G/G genotype is predominantly associated with the presence of RO, BO and OAC.
Distribution of the polymorphism in the EGF (position +61) gene demonstrated for the four study groups. The comparison of the odds ratios (OR) shows an increased risk for the development of RO, BO and OAC, with P-values of 0.006 (RO), 0.002 (BO) and 0.001 (OAC), respectively, in the homozygous G/G group compared with the healthy controls. A clear shift from the A-allele (dark gray) in the control group towards the G allele (white) in the RO, BO and OAC groups can be observed in the bars.
Risk stratification was calculated for RO, BO and OAC patients together named reflux-associated disease with the controls as a reference. The EGF A/G genotype frequency did not differ between diseased patients and controls (OR=1.05; 95% CI: 0.74–1.49). However, EGF G/G homozygosity was observed more frequently in the diseased group (OR=2.76; 95% CI: 1.45–5.24; P=0.002).
The EGF polymorphism in BO is associated with sex
The ordinal regression model was used to test for sex-specific differences in disease stage distribution between the EGF 5′ UTR genotypes.
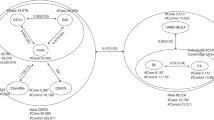
Figure 2 shows the distribution of the three genotypes for RO, BO and OAC in male (Figure 2a) and female (Figure 2b) patients. The y axis represents the chance of disease development in percentages and on the x axis the age is shown. The three genotypes are grouped per disease as lines of colored symbols. Females with RO cross all BO genotypes around the age of 80, with the G/G genotype for RO and BO crossing at younger age (RO: dark blue, BO: yellow, OAC: deep red) (Figure 2b). This suggests that females with RO and the G/G genotype have a stronger causative relation with BO than the A/A and A/G genotype. BO and the OAC genotypes never cross before the age of 80, which means that there seems no current causality between BO and OAC in females, but the causality between these diseases could appear at a higher age.
Ordinal regression model for the causality of esophageal disease in males (a) and females (b). The RO–BO–OAC cascade represents a causal relation when the lines cross. The age is shown on the x axis and the chance of developing esophageal disease is shown as percentage on the y axis. (a) Male patients develop RO from BO around the age of 60, and OAC from BO around the age of 80. The G/G genotype crosses the RO–BO line and BO–OAC line at the youngest age. (b) The genotypes for RO and BO in females cross around the age of 80 years, and the G/G genotype (RO—dark blue squares with BO—yellow squares) crosses at younger age when compared with the A/A and A/G genotype. The genotype lines for BO and OAC in females never cross.
Males with RO cross BO at the age of ∼55, and do so at a younger age than males with an A/G or A/A genotype (G/G-RO: dark blue-BO: yellow -OAC: deep red). The development of BO towards OAC appears at ∼80 years of age, with the G/G genotype crossing some years earlier than the A/A and A/G genotypes (Figure 2b). These results indicate that male RO patients with an EGF 5′ UTR G/G genotype have an increased risk for BO and OAC development, as a RO–BO–OAC disease cascade.
Figure 3 shows the distribution of the three genotypes per disease as A/A with red squares, A/G with green squares and G/G with blue squares. The y axis represents the predicted probability for disease in percentages and on the x axis the age is shown. All lines are double and to be separated in males (top) and females (bottom). The G/G genotype is associated with a higher probability for the development of RO (Figure 3a), BO (Figure 3b) and OAC (Figure 3c). Comparing all female graphs with the male graphs for BO and OAC points out that females form similar curves as males, but 20 years of age later (Figures 3a–c).
Logistic regression of the EGF (+61A/G) gene for the RO, BO and OAC development of male and female patients. The age is shown on the x axis and the predicted probability of esophageal disease is shown as percentage on the y axis. The G/G genotype (blue lines) is associated with an increased risk for RO, BO and OAC. For all instances, the upper line represents the male individuals and the lower line the female patients. Note that the lines representing female patients follow the same pattern as the male curves but seem shifted in time by ∼20 years.
EGF expression in BO in relation to genotype
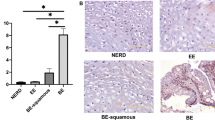
EGF and EGFR expression might influence development of RO, BO or OAC. Therefore, the variation of EGF and EGFR expression in BO biopsies per genotype was determined (Figure 4). EGF staining was observed in the cytoplasm of epithelial cells and inflammatory cells. BO biopsies varied in EGF expression (Figures 4a–c) and all OAC biopsies tested showed a strong EGF expression in the tumor (data not shown).
Typical examples of the EGF and EGFR expression. EGF was scored as 0 (negative), 1 (mild, 1–100 positive cells per microscopic field) and 2 (strong, >100 positive cells per microscopic field) on BO tissue of patients from our BO polymorphism cohort. (a) Negative staining for EGF on BO tissue. (b) Mild EGF staining (brown) of metaplastic epithelial cells and submucosal inflammatory cells is alternated by blue epithelial and inflammatory cells. (c) EGF staining is strongly present in all metaplastic epithelial cells and in most inflammatory cells. (d) Negative staining for EGFR on BO tissue. (e) EGFR staining (brown) of metaplastic epithelial cells is present in RO tissue (left) and BO tissue (right). (f) The EGFR staining is strongly present in all OAC metaplastic epithelial cells and also in BO tissue.
The EGF staining was negative in 0% of A/A, 5% of A/G and 43% of G/G patients. The positive biopsies displayed mild staining in 38% of A/A, 68% of A/G and 57% of G/G patients. Strong EGF expression was found in 62% of A/A, 27% of A/G and 0% of G/G patients. The mean EGF expression (analysis of variance, P=0.067) did not differ between the three genotype groups (A/A, A/G and G/G). However, comparisons of EGF expression among A/A versus A/G (P=0.13), A/G versus G/G (P=0.032) and A/A versus G/G (P=0.0093) genotype carriers showed that the carriage of the G allele was associated with a significantly lower local EGF expression (Figure 5a).
The median immunohistochemical EGF and EGFR scores in a subset of the BO cohort are presented in the boxplot. (a) The G/G genotype shows a significant lower local EGF expression in BO epithelium compared with the A/A and A/G carriers. The A/A genotype significantly expresses more EGF than the A/G carriers. (b) The EGFR staining is not significantly different between the three genotype groups.
EGFR expression is known to be present in (pre-) malignant tissues. Strong EGFR expression was observed in near to all biopsies of our BO cohort (28/33) (Figures 4d–f). The EGF expression was calculated for 37 biopsies, but 4 stains of the EGFR slides were unreliable and taken out of the calculations. The median EGFR expression per EGF genotype (A/A versus G/G, P=0.83) showed no significant difference (analysis of variance, P=0.094) (Figure 5b).
Discussion
Persistent severe gastro-esophageal reflux of gastric acid and bile is associated with the development of RO and BO, which are the main risk factors for developing OAC.2, 3 As EGF/EGFR has been implicated in the progression of BO into OAC,31 the EGF genotype may serve as a biomarker to assess the risk of malignant transformation in surveillance programs for patients with BO.
The EGF (+61A/G) polymorphism is associated with individual risk of esophageal, gastric, gall bladder, ovarian and renal cell cancer in a variety of studies.18, 19, 20, 21, 32 Our study shows an association between the functional EGF 5′ UTR G allele and RO-BO-OAC development.
The EGF 5′ UTR G allele frequency showed a significantly increased odds ratio of 2.6 for RO, 3.0 for BO and 4.1 for OAC, and the 5′ UTR G allele was found to be closely associated with local lower EGF expression levels in BO. The reduced EGF production in the G/G genotype could therefore mediate the development of BO and OAC. To our knowledge, we are the first to show the association of the functional EGF G/G genotype with both an enhanced chance of RO, BO and OAC development, and a decreased EGF expression in BO tissue.
A hospital-based case–control study on the association between genetic polymorphisms first needs large subgroups to show an association. For that purpose, we sampled groups of patients that by comparison with other papers are substantial, but confirmation and validation with further studies is needed.
The allele frequencies in the control cohort we used to test this polymorphism was close to equal with the frequencies reported for the European HapMap population.33 Eventually, all men at higher age develop RO, BO and OAC, as shown in our ordinal regression model. As a consequence, the control group has fewer male cases and a slightly younger age, while older males had RO complaints. The RO–BO–OAC disease cascade is thought to progress more often and faster in males than in females.34 This suggests a stronger correlation between causative agents and disease in males than in females, as we showed in the present study with the ordinal regression model.
The number of BO biopsy samples that was available in sufficient quantities for use in our immunohistochemical EGF expression study was small (n=37); we did, however, observe a significantly lower EGF expression in the G/G genotype compared with A/G or A/A carriers (P=0.0093). The precise mechanisms underlying the EGF gene interaction in BO and OAC risk are probably complex. Previous studies first showed that the studied EGF polymorphism was functional and associated with EGF serum levels. The G allele was linked to elevated EGF production in lymphocytes of malignant melanoma patients,35 and serum of patients with gastro-esophageal reflux disease.19 Second, low EGF serum levels may increase the risk of renal cell carcinoma.18 In this study, we found that there was a significantly lower EGF expression in BO cases carrying G/G homozygote than the A/A or A/G carriers.
As EGF is abundant in saliva and esophageal mucosa, serum EGF levels may not entirely reflect the endoluminal milieu of the esophagus. Indeed, a study on the local EGF expression in RO, BO and OAC indicates that EGF was depleted in RO, the EGF expression of BO epithelium was mostly superficial and EGF overexpression was observed in all histological OAC samples.24 These observations are very similar to our findings and support our hypothesis that a genetically predetermined decreased EGF expression results in an increased risk for RO, BO and OAC development.
EGFR expression was observed intracellular in RO, BO and OAC, as shown in previous studies.25 The finding of EGFR overexpression in all BO biopsies is in line with the previously reported EGFR overexpression in a wide variety of cancers of the gastrointestinal tract.36, 37, 38 High EGFR levels might well represent a negative feedback loop resulting from low EGF levels. This would suggest that elevated EGFR levels could be an indirect result of gastrointestinal neoplasia and cancer development,39 rather than a direct trigger for neoplastic development.40 That high EGFR levels not likely cause tumor development is further supported by the finding that EGFR overexpression in OAC is associated with advanced pathologic tumor classification and lymph node metastasis,28 but not with an increased risk for tumor development or survival. In line with this, a previous study showed that not EGFR but the TGF-β expression differentiated between the survival of esophageal cancer patients.41 Finally, an EGF/EGFR peptide array study demonstrated that EGF activation is significantly more prominent in squamous epithelium than in BO, concluding that EGF is an important growth factor for normal esophageal tissue regulation through binding with EGFR. Although the EGFR expression is stronger in BO compared with squamous epithelium, the activation of EGFR in BO is lower.40 This suggests that EGFR is a major receptor, but the growth factors EGF and TGF-β determine the outcome of disease. The decreased EGF activity in the esophagus could therefore trigger the promotion of a tumor environment, thereby either directly or indirectly affecting the risk for OAC development.42, 43
In conclusion, we demonstrate that the homozygous G/G genotype of the EGF 5′ UTR polymorphism is associated with a significant decrease in local EGF levels and an increased risk for the development of RO, BO and OAC. The genetically determined decreased EGF levels could result in less EGFR activation, leading to neoplastic changes, like RO, BO and OAC, but this hypothesis needs validation in other independent cohorts.
References
Blot, W. J., Devesa, S. S., Kneller, R. W. & Fraumeni Jr, J. F. Rising incidence of adenocarcinoma of the esophagus and gastric cardia. JAMA 265, 1287–1289 (1991).
Hamilton, S. R. & Smith, R. R. The relationship between columnar epithelial dysplasia and invasive adenocarcinoma arising in Barrett's esophagus. Am. J. Clin. Pathol. 87, 301–312 (1987).
Lagergren, J., Bergstrom, R., Lindgren, A. & Nyren, O. Symptomatic gastroesophageal reflux as a risk factor for esophageal adenocarcinoma. N. Engl. J. Med. 340, 825–831 (1999).
Theisen, J., Stein, H. J., Feith, M., Kauer, W. K., Dittler, H. J., Pirchi, D. et al. Preferred location for the development of esophageal adenocarcinoma within a segment of intestinal metaplasia. Surg. Endosc. 20, 235–238 (2006).
Izzo, J. G., Luthra, R., Wu, T. T., Correa, A. M., Luthra, M., Anandasabapathy, S. et al. Molecular mechanisms in Barrett's metaplasia and its progression. Semin. Oncol. 34, S2–S6 (2007).
Lembo, A., Zaman, M., Jones, M. & Talley, N. J. Influence of genetics on irritable bowel syndrome, gastro-oesophageal reflux and dyspepsia: a twin study. Aliment. Pharmacol. Ther. 25, 1343–1350 (2007).
Zheng, Z., Nordenstedt, H., Pedersen, N. L., Lagergren, J. & Ye, W. Lifestyle factors and risk for symptomatic gastroesophageal reflux in monozygotic twins. Gastroenterology 132, 87–95 (2007).
Robertson, E. V. & Jankowski, J. A. Genetics of gastroesophageal cancer: paradigms, paradoxes, and prognostic utility. Am. J. Gastroenterol. 103, 443–449 (2008).
Trudgill, N. Familial factors in the etiology of gastroesophageal reflux disease, Barrett's esophagus, and esophageal adenocarcinoma. Chest. Surg. Clin. N. Am. 12, 15–24 (2002).
Romero, Y., Cameron, A. J., Locke III, G. R., Schaid, D. J., Slezak, J. M., Branch, C. D. et al. Familial aggregation of gastroesophageal reflux in patients with Barrett's esophagus and esophageal adenocarcinoma. Gastroenterology 113, 1449–1456 (1997).
Orlando, R. C. Esophageal epithelial defense against acid injury. J. Clin. Gastroenterol. 13 (Suppl 2), S1–S5 (1991).
Orlando, R. C. Review article: oesophageal mucosal resistance. Aliment. Pharmacol. Ther. 12, 191–197 (1998).
Fujiwara, Y., Higuchi, K., Tominaga, K., Watanabe, T., Oshitani, N. & Arakawa, T. Functional oesophageal epithelial defense against acid. Inflammopharmacology 13, 1–13 (2005).
Jorissen, R. N., Walker, F., Pouliot, N., Garrett, T. P., Ward, C. W. & Burgess, A. W. Epidermal growth factor receptor: mechanisms of activation and signalling. Exp. Cell Res. 284, 31–53 (2003).
Olayioye, M. A., Neve, R. M., Lane, H. A. & Hynes, N. E. The ErbB signaling network: receptor heterodimerization in development and cancer. Embo J. 19, 3159–3167 (2000).
Laurence, D. J. & Gusterson, B. A. The epidermal growth factor. A review of structural and functional relationships in the normal organism and in cancer cells. Tumour Biol. 11, 229–261 (1990).
Singletary, S. E., Baker, F. L., Spitzer, G., Tucker, S. L., Tomasovic, B., Brock, W. A. et al. Biological effect of epidermal growth factor on the in vitro growth of human tumors. Cancer Res. 47, 403–406 (1987).
Zhu, J., Meng, X., Yan, F., Qin, C., Wang, M., Ding, Q. et al. A functional epidermal growth factor (EGF) polymorphism, EGF serum levels and renal cell carcinoma risk in a Chinese population. J. Hum. Genet. 55, 236–240 (2010).
Lanuti, M., Liu, G., Goodwin, J. M., Zhai, R., Fuchs, B. C., Asomaning, K. et al. A functional epidermal growth factor (EGF) polymorphism, EGF serum levels, and esophageal adenocarcinoma risk and outcome. Clin. Cancer Res. 14, 3216–3222 (2008).
Vishnoi, M., Pandey, S. N., Modi, D. R., Kumar, A. & Mittal, B. Genetic susceptibility of epidermal growth factor +61A>G and transforming growth factor beta1 −509C>T gene polymorphisms with gallbladder cancer. Hum. Immunol. 69, 360–367 (2008).
Araujo, A. P., Ribeiro, R., Pereira, D., Pinto, D., Sousa, B., Catarino, R. et al. Ovarian cancer and genetic susceptibility: association of A61G polymorphism in the EGF gene. Exp. Biol. Med. (Maywood) 234, 241–245 (2009).
Hamai, Y., Matsumura, S., Matsusaki, K., Kitadai, Y., Yoshida, K., Yamaguchi, Y. et al. A single nucleotide polymorphism in the 5′nslated region of the EGF gene is associated with occurrence and malignant progression of gastric cancer. Pathobiology 72, 133–138 (2005).
Upadhyay, R., Jain, M., Kumar, S., Ghoshal, U. C. & Mittal, B. Interaction of EGFR 497Arg>Lys with EGF +61A>G polymorphism: modulation of risk in esophageal cancer. Oncol. Res. 17, 167–174 (2008).
Jankowski, J., Coghill, G., Tregaskis, B., Hopwood, D. & Wormsley, K. G. Epidermal growth factor in the oesophagus. Gut 33, 1448–1453 (1992).
Jankowski, J., Murphy, S., Coghill, G., Grant, A., Wormsley, K. G., Sanders, D. S. et al. Epidermal growth factor receptors in the oesophagus. Gut 33, 439–443 (1992).
Thomas, D. M., Nasim, M. M., Gullick, W. J. & Alison, M. R. Immunoreactivity of transforming growth factor alpha in the normal adult gastrointestinal tract. Gut 33, 628–631 (1992).
Calabro, A., Orsini, B., Renzi, D., Papi, L., Surrenti, E., Amorosi, A. et al. Expression of epidermal growth factor, transforming growth factor-alpha and their receptor in the human oesophagus. Histochem. J. 29, 745–758 (1997).
Wang, K. L., Wu, T. T., Choi, I. S., Wang, H., Reseetkova, E., Correa, A. M. et al. Expression of epidermal growth factor receptor in esophageal and esophagogastric junction adenocarcinomas: association with poor outcome. Cancer 109, 658–667 (2007).
di Pietro, M. & Fitzgerald, R. C. Barrett's oesophagus: an ideal model to study cancer genetics. Hum. Genet. 126, 233–246 (2009).
Bronstein, M., Pisante, A., Yakir, B. & Darvasi, A. Type 2 diabetes susceptibility loci in the Ashkenazi Jewish population. Hum. Genet. 124, 101–104 (2008).
Li, Y., Wo, J. M., Ray, M. B., Jones, W., Su, R. R., Ellis, S. et al. Cyclooxygenase-2 and epithelial growth factor receptor up-regulation during progression of Barrett's esophagus to adenocarcinoma. World J. Gastroenterol. 12, 928–934 (2006).
Araujo, A. P., Costa, B. M., Pinto-Correia, A. L., Fragoso, M., Ferreira, P., Dinis-Ribeiro, M. et al. Association between EGF +61A/G polymorphism and gastric cancer in Caucasians. World J. Gastroenterol. 17, 488–492 (2011).
International HapMap Consortium. The International HapMap Project. Nature 426, 789–796 (2003).
van Blankenstein, M., Looman, C. W., Johnston, B. J. & Caygill, C. P. Age and sex distribution of the prevalence of Barrett's esophagus found in a primary referral endoscopy center. Am. J. Gastroenterol. 100, 568–576 (2005).
Shahbazi, M., Pravica, V., Nasreen, N., Fakhoury, H., Fryer, A. A., Strange, R. C. et al. Association between functional polymorphism in EGF gene and malignant melanoma. Lancet 359, 397–401 (2002).
Yasui, W., Hata, J., Yokozaki, H., Nakatani, H., Ochiai, A., Ito, H. et al. Interaction between epidermal growth factor and its receptor in progression of human gastric carcinoma. Int. J. Cancer 41, 211–217 (1988).
Yasui, W., Sumiyoshi, H., Hata, J., Kameda, T., Ochiai, A., Ito, H. et al. Expression of epidermal growth factor receptor in human gastric and colonic carcinomas. Cancer Res. 48, 137–141 (1988).
Tokunaga, A., Onda, M., Okuda, T., Teramoto, T., Fujita, I., Mizutani, T. et al. Clinical significance of epidermal growth factor (EGF), EGF receptor, and c-erbB-2 in human gastric cancer. Cancer 75, 1418–1425 (1995).
Tahara, E., Sumiyoshi, H., Hata, J., Yasui, W., Taniyama, K., Hayashi, T. et al. Human epidermal growth factor in gastric carcinoma as a biologic marker of high malignancy. Jpn. J. Cancer Res. 77, 145–152 (1986).
van Baal, J. W., Diks, S. H., Wanders, R. J., Rygiel, A. M., Milano, F., Joore, J. et al. Comparison of kinome profiles of Barrett's esophagus with normal squamous esophagus and normal gastric cardia. Cancer Res. 66, 11605–11612 (2006).
Iihara, K., Shiozaki, H., Tahara, H., Kobayashi, K., Inoue, M., Tamura, S. et al. Prognostic significance of transforming growth factor-alpha in human esophageal carcinoma. Implication for the autocrine proliferation. Cancer 71, 2902–2909 (1993).
Boonstra, J., Rijken, P., Humbel, B., Cremers, F., Verkleij, A. & van Bergen en Henegouwen, P. The epidermal growth factor. Cell Biol. Int. 19, 413–430 (1995).
Sethi, G., Ahn, K. S., Chaturvedi, M. M. & Aggarwal, B. B. Epidermal growth factor (EGF) activates nuclear factor-kappaB through IkappaBalpha kinase-independent but EGF receptor-kinase dependent tyrosine 42 phosphorylation of IkappaBalpha. Oncogene 26, 7324–7332 (2007).
Moons, L. M., Kusters, J. G., van Delft, J. H., Kuipers, E. J., Gottschalk, R., Geldof, H. et al. A pro-inflammatory genotype predisposes to Barrett's esophagus. Carcinogenesis 29, 926–931 (2008).
Acknowledgements
We would like to thank the participants of the pro-inflammatory genotype study44 for their materials. We also like to acknowledge Dr Honkoop and Dr Wolters for the OAC materials, Dr H Geldof and Dr WA Bode, and the nurses of the endoscopy units of the IJsselland Hospital and the Erasmus MC for their assistance in patient occlusion and collecting materials, and J Francke, A Heijens and M Ouwendijk for their assistance with the processing of the materials.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Rights and permissions
About this article
Cite this article
Menke, V., Pot, R., Moons, L. et al. Functional single-nucleotide polymorphism of epidermal growth factor is associated with the development of Barrett's esophagus and esophageal adenocarcinoma. J Hum Genet 57, 26–32 (2012). https://doi.org/10.1038/jhg.2011.124
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/jhg.2011.124
Keywords
This article is cited by
-
Genetic Biomarkers of Barrett’s Esophagus Susceptibility and Progression to Dysplasia and Cancer: A Systematic Review and Meta-Analysis
Digestive Diseases and Sciences (2016)
-
The Genetics of Barrett’s Esophagus: A Familial and Population-Based Perspective
Digestive Diseases and Sciences (2016)
-
Editorial: Genetics of Barrett's: The Bigger, the Better
American Journal of Gastroenterology (2012)