Abstract
Six human remains (dating ∼2500 years ago) were excavated from Pengyang, China, an area occupied by both ancient nomadic and farming people. The funerary objects found with these remains suggested they were nomads. To further confirm their ancestry, we analyzed both the maternal lineages and paternal lineages of the ancient DNA. From the mitochondrial DNA, six haplotypes were identified as three haplogroups: C, D4 and M10. The haplotype-sharing populations and phylogenetic analyses revealed that these individuals were closely associated with the ancient Xiongnu and modern northern Asians. Single-nucleotide polymorphism analysis of Y chromosomes from four male samples that were typed as haplogroup Q indicated that these people had originated in Siberia. These results show that these ancient people from Pengyang present a close genetic affinity to nomadic people, indicating that northern nomads had reached the Central Plain area of China nearly 2500 years ago.
Similar content being viewed by others
Introduction
The Ningxia area in China, where both ancient farming people and nomads lived 2000 years ago,1 is in the southern inner Mongolia and northern Central Plain areas (Figure 1). As the farming people migrated and expanded in the Central Plain area, they became the ancestors of the modern Han Chinese; meanwhile, the nomads occupied Siberia and Mongolia. Owing to conflicts between these two ethnic groups in the frontier, the nomads assimilated gradually into the Han Chinese population or became the minorities in northern China.1, 2 Therefore, the minorities in northern China and other northern Asians can trace their origins to the ancient nomads.1 Archeological studies of the Zhongzhuang and Wangdahu sites, located in Pengyang in the southern Ningxia area in China (Figure 1), indicated that they were occupied from ∼770 BC to 256 BC during the Dongzhou period. The funerary objects included many skulls of animals, bronze swords and daggers, but no farming tools. The ancient Pengyang people were identified anthropologically as northern Asians, implying that they were nomads. In this study, we analyzed the maternal and paternal lineages of the ancient DNA from the remains, confirming the evolution of the populations living in this area.
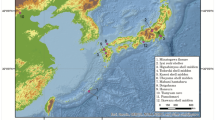
Geographic distribution of the ancient individuals from Pengyang and their sharing populations. (a) Geographic location of Pengyang. (b) Distribution of the populations shared by PZ1, PZ2, PW4, PW6 and PW7. (c) Distribution of the populations shared by PW5. Every dot represents an individual. Only the last three digits are given, for example, site 223 represents 16 223.
Materials and methods
Fragments (∼2 cm long) of the unbroken femurs were sampled from six well-preserved ancient human remains at two sites, Zhongzhuang and Wangdahu (Table 1). All analyses were conducted in the professional ancient DNA laboratory, following the criteria for ancient DNA studies.3 Ensuring authenticity, DNA extraction and polymerase chain reaction protocols were previously published.4 The mitochondrial hypervariable I segments (16 050–16 409) were amplified using three overlapping primer pairs. The mitochondrial haplogroups were identified using the amplified product-length polymorphism method.5 The sex of each sample was determined by the amelogenin gene polymerase chain reaction test. Y chromosome single-nucleotide polymorphisms M175 and M242 were determined by sequencing to type the haplogroups O and Q.6 In the Y chromosome single-nucleotide polymorphism procedure, any male researcher was excluded and the ancient female individuals were also amplified as negative controls. All primers in this study are listed in Supplementary Table S1. The sharing populations were searched from the published data of modern people across East Asia and Siberia, the source of which is provided in Supplementary Table S2. The mitochondrial hypervariable I sequence of every researcher was obtained to identify potential laboratory-based contamination (Supplementary Table S3).
Results and discussion
DNA samples from all individuals were successfully amplified, and the obtained mitochondrial hypervariable I sequences were inferred to be authentic. We found six haplotypes from haplogroups C, D4 and M10 (Table 1). Haplogroup D4 has a high distribution frequency (8–22.3%) in northern Asians7, 8 and eastern Asians.9 Haplogroup C has a very high distribution frequency (15.9–67.2%) in Siberia,7 but a low frequency in China (3.5%).9 Haplogroup M10 is a new sub-haplogroup of M10 with a frequency range of 0–2.3% in modern Han Chinese,11 while a few M10 individuals have also been found among Mongolians and Yakuts.8, 12
A phylogenetic tree was constructed to determine the genetic relationship between the ancient Pengyang people and related populations (Figure 2). In haplogroup C, the Han Chinese originating from the farming people1, 9, 10 are concentrated in one group. Meanwhile, PW4 is clustered into another group with minorities in northern China, northern Asians and ancient nomads,2, 7, 12, 13, 14, 15, 16, 17, 18, 19 suggesting that PW4 is closely related to the nomads. In haplogroups D4 and M10, the ancient people from Pengyang consistently occupy the same nodes as northern Asians, northern minorities of China and ancient nomads, showing a close genetic relationship among them.
The maximum parsimony trees of median-joining networks of the mtDNA haplogroups of Pengyang ancient people in comparison with other populations. A phylogenetic tree was constructed by Network software ver. 4.5.1.0 (http://www.fluxus-engineering.com) using maximum parsimony methods and the mtDNA data of northern Han Chinese,9, 10 northern Asians,7, 12, 13, 14 minorities in northern China2, 15 and the ancient nomadic people.16, 17, 18, 19
To further understand the genetic relationship among these populations, the six haplotypes from our study were used as queries to search for sharing populations in the published data of modern people across East Asia and Siberia, and we selected the matched sequences that belong to the same haplogroups for further comparisons (Figure 1; Supplementary Table S2). PW4 (16 171–16 223–16 298–16 327–16 344–16 357) shares a haplotype with 58 individuals. Among these, 49 individuals are from outer Mongolia and Siberia, 8 are from Uighur of Kazakstan and minorities of northern China who also originated from northern Asia,1, 2 and only 1 is from the Daic population in southern China. PZ2 (16 082–16 223–16 362) shares a haplotype with four Siberians and two mongolians. PW6 (16 223–16 287–16362) shares a haplotype with only one Kazak of China. Interestingly, all individuals sharing haplotypes with PZ2 and PW6 originated from nomads. PW7 (16 093–16 129–16 193–16 223–16 311–16 357) shares a haplotype with seven people, one of whom is an inner Mongolian, but the origins of the other six individuals do not trace back to the nomads. Haplotype 16 223–16 311–16 362 of PZ1 is observed in 25 people, including three Buryats, one Mongolian and five Tibetans, whose origin can be traced back to the nomads.1 Moreover, this haplotype is also found in ancient Xiongnus from northern Asia.16 Therefore, it is possible that PZ1 originated from the nomads. Haplotype 16 223–16 362 of PW5 is an ancient haplotype in haplogroup D and is found in various populations. The haplotype 16 223–16 362 is highly prevalent in eastern Asians, but its frequency is also high in northern Asians (Figure 1) and in ancient Xiongnus.16 Overall, every sample from Pengyang tested is related to northern Asians or northern minorities of China, and bears a high resemblance to northern nomads.
By surveying Y chromosome single-nucleotide polymorphisms, all four male individuals were typed as haplogroup Q, believed to have arisen in Siberia (Table 1). Generally, haplogroup Q is distributed at a very high frequency throughout Siberia, but at low frequencies outside of Siberia.20 The frequency of haplogroup Q among Han Chinese was reported as 2.8%, whereas haplogroup O is predominant at 66.8%.9 These results demonstrate a close patrilineal genetic relationship between the ancient Pengyang people and the nomads.
Historical records documented that the nomadic culture was predominant in southern Inner Mongolia in the Dongzhou period, which was associated with historical events such as climate cooling, grassland degradation and farming people losing control of the border area.1 Taken together, this study suggests that the northern nomads may have reached the southern Ningxia area in the Dongzhou period.
References
Du, R. & Yip, V. F. (eds). Ethnic Groups in China. (Science Press, Beijing, 1993).
Kong, Q. P., Yao, Y. G., Liu, M., Shen, S. P., Chen, C., Zhu, C. L. et al. Mitochondrial DNA sequence polymorphisms of five ethnic populations from northern China. Hum. Genet. 113, 391–405 (2003).
Cooper, A. & Poinar, H. N. Ancient DNA: do it right or not at all. Science 289, 1139 (2000).
Xie, C. Z., Li, C. X., Cui, Y. Q., Zhang, Q. C., Fu, Y. Q., Zhu, H. et al. Evidence of ancient DNA reveals the first European lineage in Iron Age Central China. Proc. Biol. Sci. 274, 1597–1601 (2007).
Umetsu, K., Tanaka, M., Yuasa, I., Saitou, N., Takeyasu, I., Fuku, N. et al. Multiplex amplified product-length polymorphism analysis for rapid detection of human mitochondrial DNA variations. Electrophoresis 22, 3533–3582 (2001).
Karafet, T. M., Mendez, F. L., Meilerman, M. B., Underhill, P. A., Zegura, S. L. & Hammer, M. F. New binary polymorphisms reshape and increase resolution of the human Y chromosomal haplogroup tree. Genome Res. 18, 830–838 (2008).
Starikovskaya, E. B., Sukernik, R. I., Derbeneva, O. A., Volodko, N. V., Ruiz-Pesini, E., Torroni, A. et al. Mitochondrial DNA diversity in indigenous populations of the southern extent of Siberia, and the origins of Native American haplogroups. Ann. Hum. Genet. 69, 67–89 (2005).
Kolman, C. J., Sambuughin, N. & Bermingham, E. Mitochondrial DNA analysis of Mongolian populations and implications for the origin of New World founders. Genetics 142, 1321–1334 (1996).
Wen, B., Li, H., Lu, D., Song, X., Zhang, F., He, Y. et al. Genetic evidence supports demic diffusion of Han culture. Nature 431, 302–305 (2004).
Yao, Y. G., Kong, Q. P., Bandelt, H. J., Kivisild, T. & Zhang, Y. P. Phylogeographic differentiation of mitochondrial DNA in Han Chinese. Am. J. Hum. Genet. 70, 635–651 (2002).
Gao, S. Z., Yang, Y. D., Xu, Y., Zhang, Q. C., Zhu, H. & Zhou, H. Tracing the genetic history of the Chinese people: mitochondrial DNA analysis of aneolithic population from the Lajia site. Am. J. Physiol. Anthropol. 133, 1128–1136 (2007).
Bermisheva, M. A., Kutuev, I. A., Spitsyn, V. A., Villems, R., Batyrova, A. Z., Korshunova, T. et al. Analysis of mitochondrial DNA variation in the population of Oroks. Genetika 41, 78–84 (2005).
Pakendorf, B., Wiebe, V., Tarskaia, L. A., Spitsyn, V. A., Soodyall, H., Rodewald, A. et al. Mitochondrial DNA evidence for admixed origins of central Siberian populations. Am. J. Physiol. Anthropol. 120, 211–224 (2003).
Volodko, N. V., Starikovskaya, E. B., Mazunin, I. O., Eltsov, N. P., Naidenko, P. V., Wallace, D. C. et al. Mitochondrial genome diversity in arctic Siberians, with particular reference to the evolutionary history of Beringia and Pleistocenic peopling of the Americas. Am. J. Hum. Genet. 82, 1084–1100 (2008).
Yao, Y. G., Kong, Q. P., Wang, C. Y., Zhu, C. L. & Zhang, Y. P. Different matrilineal contributions to genetic structure of ethnic groups in the silk road region in china. Mol. Biol. Evol. 21, 2265–2280 (2004).
Keyser-Tracqui, C., Crubezy, E. & Ludes, B. Nuclear and mitochondrial DNA analysis of a 2,000-year-old necropolis in the Egyin Gol Valley of Mongolia. Am. J. Hum. Genet. 73, 247–260 (2003).
Yu, C., Xie, L., Zhang, X., Zhou, H. & Zhu, H. Genetic analysis on Tuoba Xianbei remains excavated from Qilang Mountain Cemetery in Qahar Right Wing Middle Banner of Inner Mongolia. FEBS Lett. 580, 6242–6246 (2006).
Wang, H., Ge, B., Mair, V. H., Cai, D., Xie, C., Zhang, Q. et al. Molecular genetic analysis of remains from Lamadong cemetery, Liaoning, China. Am. J. Phys. Anthropol. 134, 404–411 (2007).
Fu, Y., Zhao, H., Cui, Y., Zhang, Q., Xu, X., Zhou, H. et al. Molecular genetic analysis of Wanggu remains, Inner Mongolia, China. Am. J. Phys. Anthropol. 132, 285–291 (2007).
Zegura, S. L., Karafet, T. M., Zhivotovsky, L. A. Hammer, M. F. High-resolution SNPs and microsatellite haplotypes point to a single, recent entry of Native American Y chromosomes into the Americas. Mol. Biol. Evol. 21, 164–175 (2004).
Acknowledgements
This work was supported by a grant from the Key Laboratory for Evolution of Past Life and Environment in Northeast Asia, Ministry of Education, China (EPLENEA). We are grateful to the Institute of Cultural and Historical Relics and Archaeology in the Ningxia Hui Nationality Autonomous Region, China, for providing the samples. We also thank Wenxia Song from the Department of Cell Biology and Molecular Genetics of Maryland University for assistance with revision of the paper.
Author information
Authors and Affiliations
Corresponding author
Additional information
Supplementary Information accompanies the paper on Journal of Human Genetics website
Supplementary information
Rights and permissions
About this article
Cite this article
Zhao, YB., Li, HJ., Cai, DW. et al. Ancient DNA from nomads in 2500-year-old archeological sites of Pengyang, China. J Hum Genet 55, 215–218 (2010). https://doi.org/10.1038/jhg.2010.8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/jhg.2010.8
Keywords
This article is cited by
-
Origin of ethnic groups, linguistic families, and civilizations in China viewed from the Y chromosome
Molecular Genetics and Genomics (2021)
-
Genetic analysis of male Hungarian Conquerors: European and Asian paternal lineages of the conquering Hungarian tribes
Archaeological and Anthropological Sciences (2020)
-
Dispersals of the Siberian Y-chromosome haplogroup Q in Eurasia
Molecular Genetics and Genomics (2018)