Abstract
Coronary atherosclerosis is a complex and progressive condition that involves many biological pathways, including the oxidative stress and inflammatory response pathways. To investigate the association between common genetic variation within these two pathways and coronary atherosclerosis, we performed a comprehensive two-stage candidate gene association study in a Chinese Han population. In stage I, 936 tag single-nucleotide polymorphisms (SNPs) within 116 candidate genes were genotyped in 293 coronary atherosclerosis cases and 293 age- and gender-matched healthy controls. In stage II, 51 SNPs from stage I were selected and further genotyped in an additional 1030 cases and 764 controls. In allele- and genotype-based association tests of stage II and a meta-analysis across the two stages, we identified three SNPs within three genes significantly associated with the disease, namely rs3212556 in ITGA2 (PCMH=9.20 × 10−5), rs854563 in PON1 (PCMH=1.92 × 10−4) and rs9283851 in THBS2 (PCMH=3.00 × 10−3). Haplotype analysis provided further supporting evidence for the association of rs3212556 (Pglobal<10−4) and rs854563 (Pglobal<10−4). Our study has identified three SNPs within ITGA2, PON1 and THBS2 that are associated with coronary atherosclerosis.
Similar content being viewed by others
Introduction
Cardiovascular disease is the leading cause of illness and death in developed countries and some developing countries, and has become a significant health problem worldwide.1, 2 The primary cause of cardiovascular disease is atherosclerosis, which leads to ischemia of the heart, brain or extremities resulting in infarction. Coronary atherosclerosis is a progressive disease that involves multiple processes, including imbalance of reactive oxygen species,3 diapedesis of monocytes across the endothelial barrier, activation of neutrophils, T lymphocytes, macrophage cells and platelets, upregulation of cell adhesion molecules, release of various cytokines and chemokines, proliferation of smooth muscle cells4 and matrix alterations.5 There is substantial evidence to suggest that two biological pathways, namely the oxidative stress and inflammatory response pathways, have important roles in the pathogenesis of atherosclerosis. Reactive oxygen species are produced as a by-product of aerobic respiration and substrate oxidation. The levels of these free radicals in body tissues are regulated by a defense system composed of many enzymes and nonenzymatic antioxidants. High doses and/or inadequate removal of reactive oxygen species result in oxidative stress, which may cause severe metabolic malfunctions and damage to biological macromolecules. This subsequently contributes to many human diseases, including cardiovascular disease, cancer and immune disorders. Several studies have suggested that components of this pathway, including superoxide dismutase, glutathione peroxidase, bilirubin and paraoxonase, influence the development of coronary artery disease (CAD).3 Furthermore, factors involved in inflammatory response are recognized to be crucial in the development of atheromatous plaques, thrombosis and atherogenesis.4 An increasing number of studies have suggested that atherosclerosis is an inflammatory disorder, in which immune mechanisms interact with metabolic risk factors to initiate, propagate and activate lesions in the arterial tree.6, 7
Epidemiological studies have suggested many important genetic and environmental risk factors for atherosclerosis.8 Recent genome-wide association studies of CAD have led to the identification of genetic variants and chromosome regions (namely 9p21.3, 1q41), which are significantly and reproducibly associated with this disease.9 In addition, several candidate gene studies have been conducted, testing variants in genes involved in inflammation, for example, CD14, TLR4 and VCAM1, for association with atherosclerosis and significant associations have been reported by some groups.10, 11 Notably, the majority of these studies were conducted in European populations and the results could not be replicated in Chinese Han populations. This suggests that there are substantial differences in the genetic contributors to CAD between these populations.12, 13
In this study, we performed a pathway-based candidate gene study of coronary atherosclerosis using a tag single-nucleotide polymorphism (SNP) approach for interrogating common genetic variation within the candidate genes of interests. The study was conducted in two stages. In the stage I discovery study, tag SNPs from all candidate genes of interest were genotyped in a well-matched case–control sample. In the stage validation II study, tag SNPs showing suggestive association in the stage I study were selected and genotyped in an independent and larger case–control sample to test for replication. By using such a two-stage design using independent discovery and validation samples, we have attempted not only to increase the efficiency of the association analysis but also to identify true genetic risk factors for coronary atherosclerosis in the Chinese Han population using stringent criteria for declaring association.
Materials and methods
Study sample
We collected atherosclerosis cases and normal controls on the basis of diagnosis by coronary angiography from the cardiovascular care units of three hospitals in Shanghai. All participants were of the Han Chinese origin from Shanghai and the neighboring provinces. The samples used in the stage I study included 293 cases with at least 50% atherosclerosis occlusion in the above two branches of the coronary artery and 293 age- and gender-matched normal controls without coronary artery stenosis. The stage II study included an additional 1030 cases with coronary atherosclerosis and 764 normal controls. At enrolment, anthropometric measures, medication usage and family history data were collected from each subject by a trained interviewer. The demographic and risk factor information of all case and control samples are summarized in Table 1. All the participants gave informed consent, and the study protocol was approved by the Ethics Review Committee of the Chinese National Human Genome Center at Shanghai. Genomic DNA of all samples was isolated from whole blood using FlexiGene DNA Kit (Qiagen, Valencia, CA, USA).
Genes and SNP marker selection
Our current study focused on two biological processes involved in coronary atherosclerosis, the antioxidative pathway and the inflammatory pathway. By an extensive review of literature and the information at the Gene Ontology database (http://geneontology.org/), we identified 116 candidate genes of the two pathways that are likely involved in the pathogenesis of coronary atherosclerosis. Of the 116 selected candidate genes, 32 are from the antioxidative pathway and 84 are from the inflammatory pathway. To interrogate common genetic variants within these candidate genes, we used diverse approaches for identifying suitable tag SNPs. In all, 27 of the 116 candidate genes were formerly studied by extensive resequencing. Tag SNPs within these 27 genes were selected on the basis of the genetic variation information from the resequencing database (EGP/PGA) (http://pga.gs.washington.edu/VG2.html) using a haplotype-tagging approach and a selection algorithm similar to that used in the study by Carlson et al.,14 with criterion of r2>0.8 and minor allele frequency (MAF)>0.05. For the second group of 66 candidate genes, tag SNPs were selected on the basis of the HapMap Chinese Han data (phase II), such that all common genetic variants (MAF>0.05) within these genes could be captured by the tag SNPs at r2>0.8 (http://www.hapmap.org). For the remaining third group of 23 candidate genes that were not resequenced, we selected SNPs that were uniformly distributed (the marker density reaches 2–3 kb for r2>0.8 in the EGP database), informative (heterozygosity >5%) and potentially functional (coding nonsynonymous and promoter SNPs) in dbSNP (Build 118). In total, 267 SNPs from the 32 genes of the antioxidative pathway and 669 SNPs from the 84 genes of the inflammatory pathway were selected and genotyped in 293 cases and 293 matched controls in the stage I study (Supplementary Data 1). In the stage II validation study, we selected 54 out of the 58 SNPs (excluding 4 SNPs in high linkage disequilibrium (LD) r2=1 with existing SNPs) that showed association at the significance level of 0.05 in the phase I study and genotyped them in an additional 1030 cases and 764 controls (Supplementary Data 2).
Genotyping and quality control
SNPs genotyping was performed on BeadLab SNP Genotyping System (Illumina, San Diego, CA, USA), combining high-density oligonucleotide array and multiplex thermocycled primer extension, and GenomeLab SNPstream Genotyping System (Beckman Coulter, Brea, CA, USA) combining fluorescent minisequencing and multiplex tag-array. All SNPs in phase I and phase II were partitioned into two Illumina oligonucleotide primer sets and several 12-plex and 48-plex panels by the GenomeLab SNPstream Genotyping System. To check the reliability and reproducibility of the genotyping quality, we placed one negative control (water) and three DNA samples in duplicates on every 96-well DNA plate. All genotyping data with low signal-to-noise ratio and poor genotyping clusters were excluded from further analysis. Finally, 894 SNPs in phase I and 51 SNPs in phase II with MAF>0.01 and call rate >0.9 were successfully genotyped and used for the association analysis. The overall genotype call rate was 98.8%, and the reproducibility rate was >99%.
Statistical methods
The genotypes were tested for Hardy–Weinberg equilibrium (HWE) using Fisher’s exact test15 in cases and controls separately. In the stage I discovery study, single-marker association analysis was performed under allelic model using Fisher's exact test. A low significance threshold of P=0.05 was used to select SNPs for the stage II validation analysis to maximize the power of study; at the current sample size (293 cases and 293 controls) and significance threshold, we estimate that we have >80% power to detect a common risk allele (MAF 20%) with an odds ratio (OR) of ⩾1.5. In the stage II validation study, single-marker association analysis was performed under allelic models in the validation samples. For the positive SNPs (P<0.05) in phase II, we performed a meta-analysis across the phase I and phase II sample sets. A Mantel–Haenszel common OR and a Fisher's combined P-value were calculated across the two sample sets. For the top SNPs identified by the meta-analysis, genotype-based association under dominant, recessive and additive models was performed using logistic regressions. We also performed the association analysis adjusting for gender, hypertension, diabetes, smoking, drinking, triglycerides, total cholesterol, high-density lipoprotein and low-density lipoprotein levels in all samples of stages I and II using logistic regressions. Haplotype analysis was performed in the combined phase I and II samples. LD structure was examined using Haploview,16 and LD blocks were defined using pairwise D′ values all >0.90 within the region. Haplotype analyses were carried out using Haplo.stats v1.2.2 (Mayo Clinic, Scottsdale, AZ, USA).17 Differences in haplotype frequencies between cases and controls were tested using a score test, and OR was calculated with the most common haplotype as the reference. The global P-value for each haplotype block was given on the basis of the global score statistic and was calculated through 10 000 simulations.
URLs. The URLS used were as follows: Haploview: http://www.broad.mit.edu/mpg/haploview/index.php and Haplo.stats: http://www.mayo.edu/hsr/people/schaid.html.
Results
Three SNPs associated with coronary atherosclerosis in the Chinese Han population
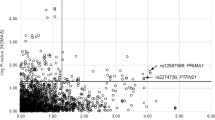
In the stage I study, the single-marker association analysis of 894 tag SNPs from 116 genes was performed using allele-based tests in 293 cases and 293 age- and gender-matched controls. A total of 58 SNPs from 21 genes, including CD36, ITGA2, VCAM1, THBS2 showed association at a significance level of P=0.05. The genotype distributions of the 58 SNPs in control samples are all under HWE (PHWE<0.05). The strongest association was observed at rs2071303 within the HFE gene (Pallelic=4.37 × 10−4) (Supplementary Data; Supplementary Table 1). Nine SNPs within ITGA2 showed association with coronary atherosclerosis. However, none of the identified associations survived the Bonferroni correction for multiple testing (P<5.59 × 10−5). A total of 51 SNPs from the stage I study were analyzed in our stage II validation study in which an additional 1030 cases and 764 controls were genotyped successfully (Supplementary Table 2). Of the 51 SNPs, 3 SNPs within three genes, rs3212556 in ITGA2 (Pallelic=1.96 × 10−3), rs854563 in PON1 (Pallelic=2.57 × 10−3) and rs9283851 in THBS2 (Pallelic=0.034), showed association with atherosclerosis in the validation samples. The genotype distributions of these three SNPs are under HWE in control samples (PHWE>0.05). A meta-analysis of the combined stage I and II data set using the Mantel–Haenszel method revealed stronger associations at the three SNPs, rs3212556 (OR=0.76, 95% confidence interval (95% CI), 0.66–0.87, P=9.20 × 10−05), rs854563 (OR=0.55, 95% CI, 0.4–0.75, P=1.92 × 10−04) and rs9283851 (OR=0.74, 95% CI, 0.6–0.9, P=3.00 × 10−03). On the basis of the frequency of the risk allele and the estimated effect size, our study had 100% power to detect rs3212556 and rs9283851 and only 45% power to detect rs854563 at a significance level of 0.05. These SNPs were associated with similar effect sizes in the two data sets, with each showing a clear dosage effect in the genotypic tests and hence best fitting an additive model of association. To evaluate the impact of known risk factors on the identified genetic association at three SNPs, we conducted an association analysis in stage I and II samples with adjustment for the effects of the known risk factors for coronary heart disease, including gender, hypertension status, diabetes status, triglycerides, total cholesterol, high-density lipoprotein and low-density lipoprotein levels and found that the association remained significant even after the adjustment (rs3212556 Padjusted=3.44 × 10−3, rs854563 Padjusted=4.72 × 10−3, rs9283851 Padjusted=2.44 × 10−4) (Table 2).
Two haplotypes in ITGA2 and PON1 associated with the disease
In addition to the single-marker association analyses, we performed haplotype association analysis in the combined stage I and II samples. We analyzed the LD patterns within the three genes, ITGA, THBS2 and PON1. We identified three haplotype blocks within ITGA2 and PON1 in which pairwise D′ values between SNPs within one block were all >0.90, whereas THBS2 was tagged by a single SNP rs9283851. We then performed haplotype association analysis within each of the three LD blocks. The haplotype analysis identified association in two haplotype blocks within ITGA2 (Pglobal<10−4) and PON1 (Pglobal<10−4) (Table 3). For the identified LD block (rs3212430–rs3212433–rs3212435–rs3212436–rs2287950–rs3212556) within ITGA2, the major haplotype (C-C-G-G-C-A) showed a significant risk effect, whereas the haplotype (T-T-G-C-C-A) showed protective effect (OR=0.18, 95% CI, 0.09–0.38, P<10−4). The result of the haplotype analysis is consistent with the single-SNP association results at rs3212556. For LD block (rs854560–rs705378–rs854563) in PON1, the haplotype (T-T-A) also showed significant protective effect (OR=0.59, 95% CI, 0.42–0.81, P=1.1 × 10−3).
Discussion
To our knowledge, this is the first study to explore the association between coronary atherosclerosis and genetic variants in two biological pathways, response to oxidative stress and inflammatory process, in the Chinese Han population. All cases and controls in this study were recruited from the same geographic region (central) of China. Although the absence of genome-wide data in this candidate gene study makes it difficult to control for possible effects of population stratification, previous work has shown that geographic matching in the Chinese Han populations is a good proxy for genetic matching;18 hence, we expect the genetic background of cases and controls to be well matched. Moreover, to avoid false-positive associations caused by differences in age, gender and other covariates, we selected cases and controls that are well matched for age and gender, and ensured that the associations remained significant after adjustment for several known risk factors for CAD.
We provided evidence that the common genetic variation within ITGA2 was associated with the risk for coronary atherosclerosis in the Chinese Han population. The single-SNP analysis identified significant association at rs3212556 located within the intron 15 of ITGA2 (PCMH=9.20 × 10−05). The association shows a clear dosage effect with OR het=0.89 (95% CI, 0.73–1.09) and OR hom=0.45 (95% CI, 0.31–0.65). The association was also supported by the haplotype analysis in which a haplotype composed of six SNPs within ITGA2, including rs3212556, showed strong association with coronary atherosclerosis. The risk-associated haplotype covers the region of intron 2 to intron 15 of ITGA2. In dbSNP, there are 14 coding SNPs within this region. Four coding polymorphisms in this region (807C>T, 837G>A 873A>G and 1648G>A) have been reported to be associated with the density of integrin α2β1 receptor in human platelets.19, 20 Previous studies have also indicated that the α2 integrin subunit is involved in monocyte attachment to collagen IV.21 Therefore, we speculate that the ITGA2 rs3212556− A allele may increase the risk for coronary atherosclerosis by changing the density of integrin α2β1 receptor. Further functional studies will be required to prove this hypothesis on the pathogenesis of atherosclerosis in Chinese populations, and to identify the true functional variant among tightly linked SNPs within the LD block.
Our study also provided evidence for the association of PON1 with atherosclerosis risk. The relationship between many enzymatic and nonenzymatic antioxidants and coronary heart disease has been investigated in many studies. Several studies have reported that genetic variation of PON1 influences the activity of paraoxonase in atherosclerosis, and some coding SNPs such as Q192R (rs662) and L55M (rs854560) were shown to be associated with CAD. However, these findings were not always consistent.22, 23 In our study, we did not replicate any of the reported associations, but instead identified association at rs854563 (PCMH=1.92 × 10−04), which lies within the same LD block as the L55M variant. rs854563 showed stronger association than the L55M variant itself or the haplotype tagged by both SNPs. Further work will be required to identify the true functional variant. The role of PON1 in CAD has been extensively debated but remains unresolved. A study by Mackness et al.24 has shown that PON1 activity and concentration may be important in the development of CAD. Genetic factors may influence the activity of paraoxonase, which may be more critical than its concentration in atherosclerosis.25
In addition to ITGA2 and PON1, our study suggested an association within THBS2. In previous studies, THBS2 has consistently been suggested to have a genetic role in atherosclerosis.26, 27 Thrombospondins constitute a family of extracellular matrix glycoproteins, of which THBS2 is implicated in adapting and modulating cell–matrix interactions by interacting with cell-surface receptors, cytokines, growth factors, proteases and structural proteins.28 THBS2 may influence CAD risk through its participation in the regulation of matrix metalloproteinase-2, a protein linked to the vulnerability of atherosclerotic plaque. THBS2-null fibroblasts produce twofold larger amounts of this protein,29 which was shown to be lower in CAD patients than in controls.30 Alternatively, THBS2-absent mice have an increased vascular density and a bleeding tendency, both of which have been hypothesized to reduce the risk of atherosclerosis.31 Our confirmed association of THBS2 with CAD warrants further functional study of rs9283851 and other polymorphisms in proximity.
The strength of this study is reflected by the comprehensive investigation of the candidate genes of two important biological processes known to be involved in the pathogenesis of coronary atherosclerosis. Another noteworthy aspect lies in conducting the study in the Chinese Han population that has been greatly underrepresented in earlier genetic association studies of coronary atherosclerosis. However, we recognize that this study has a relatively small sample size and thus limited power for identifying genetic risk variants, especially those with moderate genetic effects and/or low population frequency. For example, in our stage I discovery study with 293 cases and 293 controls, we estimate that we had ∼85% power to detect a risk allele of MAF 20% and an OR of 1.5 (α=0.05), but only ∼27% power to detect one with an OR of 1.2 (Genetic Power Calculator; S. Purcell & P. Sham, UK. http://pngu.mgh.harvard.edu/~purcell/gpc/). This may account for the failure of the associations identified to survive the stringent Bonferroni correction for multiple testing. Nonetheless, by using a two-stage design and validating the significant associations from stage 1 in an independent stage 2 study with a larger sample size, we sought to increase the stringency for declaring significant association and thus to minimize the chance of false-positive associations.
In summary, we have shown that three common genetic variants within ITGA2, PON1 and THBS2 are associated with coronary atherosclerosis in Chinese Han population. Additional validation studies with larger sample sizes will be required to further confirm these associations and to discover new ones.
References
WHO publishes definitive atlas on global heart disease and stroke epidemic. Indian J. Med. Sci 58, 405–406 (2004).
Reddy, K. S. Cardiovascular disease in non-Western countries. N. Engl. J. Med. 350, 2438–2440 (2004).
Mates, J. M., Perez-Gomez, C. & Nunez de Castro, I. Antioxidant enzymes and human diseases. Clin. Biochem. 32, 595–603 (1999).
Ross, R. Atherosclerosis–an inflammatory disease. N. Engl. J. Med. 340, 115–126 (1999).
Dzau, V. J., Braun-Dullaeus, R. C. & Sedding, D. G. Vascular proliferation and atherosclerosis: new perspectives and therapeutic strategies. Nat. Med. 8, 1249–1256 (2002).
Hansson, G. K. Inflammation, atherosclerosis, and coronary artery disease. N. Engl. J. Med. 352, 1685–1695 (2005).
Libby, P. Inflammation in atherosclerosis. Nature 420, 868–874 (2002).
Lusis, A. J. Atherosclerosis. Nature 407, 233–241 (2000).
Wellcome Trust Case Control Consortium. Genome-wide association study of 14 000 cases of seven common diseases and 3000 shared controls. Nature 447, 661–678 (2007).
Arroyo-Espliguero, R., Avanzas, P., Jeffery, S. & Kaski, J. C. CD14 and toll-like receptor 4: a link between infection and acute coronary events? Heart 90, 983–988 (2004).
Edfeldt, K., Bennet, A. M., Eriksson, P., Frostegard, J., Wiman, B., Hamsten, A. et al. Association of hypo-responsive toll-like receptor 4 variants with risk of myocardial infarction. Eur. Heart J. 25, 1447–1453 (2004).
McDermott, D. H., Yang, Q., Kathiresan, S., Cupples, L. A., Massaro, J. M., Keaney, J. F. Jr et al. CCL2 polymorphisms are associated with serum monocyte chemoattractant protein-1 levels and myocardial infarction in the Framingham. Heart Study Circ. 112, 1113–1120 (2005).
Zhang, X., Han, Y., Kang, J. & Yan, C. A monocyte chemoattractant protein-1 gene polymorphism is not associated with coronary artery disease in a Han Chinese population. Clin. Chim. Acta. 403, 241–243 (2009).
Carlson, C. S., Eberle, M. A., Rieder, M. J., Yi, Q., Kruglyak, L. & Nickerson, D. A. Selecting a maximally informative set of single-nucleotide polymorphisms for association analyses using linkage disequilibrium. Am. J. Hum. Genet. 74, 106–120 (2004).
Hudson, R. R. Genetic Data Analysis. Methods for Discrete Population Genetic Data, Bruce S. Weir. Sinauer, Sunderland, MA, 1990. xiv, 377 pp., illus. $48; paper, $27. Science. 250, 575 (1990).
Barrett, J. C., Fry, B., Maller, J. & Daly, M. J. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics 21, 263–265 (2005).
Schaid, D. J., Rowland, C. M., Tines, D. E., Jacobson, R. M. & Poland, G. A. Score tests for association between traits and haplotypes when linkage phase is ambiguous. Am. J. Hum. Genet. 70, 425–434 (2002).
Chen, J., Zheng, H., Bei, J. X., Sun, L., Jia, W. H., Li, T. et al. Genetic structure of the Han Chinese population revealed by genome-wide SNP variation. Am. J. Hum. Genet. 85, 775–785 (2009).
Kritzik, M., Savage, B., Nugent, D. J., Santoso, S., Ruggeri, Z. M. & Kunicki, T. J. Nucleotide polymorphisms in the alpha 2 gene define multiple alleles that are associated with differences in platelet alpha 2beta 1 density. Blood 92, 2382–2388 (1998).
Ajzenberg, N., Berroeta, C., Philip, I., Grandchamp, B., Ducellier, P., Huart, V. et al. Association of the -92C/G and 807C/T polymorphisms of the {alpha}2 subunit gene with human platelets {alpha}2{beta}1 receptor density. Arterioscler. Thromb. Vasc. Biol. 25, 1756–1760 (2005).
Kostidou, E., Koliakos, G., Paletas, K. & Kaloyianni, M. Monocyte attachment and migration through collagen IV in diabetes mellitus. Mol. Cells 25, 452–456 (2008).
Serrato, M. & Marian, A. J. A variant of human paraoxonase/arylesterase (HUMPONA) gene is a risk factor for coronary artery disease. J. Clin. Invest. 96, 3005–3008 (1995).
Christiansen, L., Bathum, L., Frederiksen, H. & Christensen, K. Paraoxonase 1 polymorphisms and survival. Eur. J. Hum. Genet. 12, 843–847 (2004).
Mackness, B., Davies, G. K., Turkie, W., Lee, E., Roberts, D. H., Hill, E. et al. Paraoxonase status in coronary heart disease: are activity and concentration more important than genotype? Arterioscler. Thromb. Vasc. Biol. 21, 1451–1457 (2001).
Mackness, B., Durrington, P., McElduff, P., Yarnell, J., Azam, N., Watt, M. et al. Low paraoxonase activity predicts coronary events in the Caerphilly Prospective Study. Circulation 107, 2775–2779 (2003).
McCarthy, J. J., Parker, A., Salem, R., Moliterno, D. J., Wang, Q., Plow, E. F. et al. Large scale association analysis for identification of genes underlying premature coronary heart disease: cumulative perspective from analysis of 111 candidate genes. J. Med. Genet. 41, 334–341 (2004).
Boekholdt, S. M., Trip, M. D., Peters, R. J. G., Engelen, M., Boer, J. M. A., Feskens, E. J. M. et al. Thrombospondin-2 polymorphism is associated with a reduced risk of premature myocardial infarction. Arterioscler. Thromb. Vasc. Biol. 22, e24–27 (2002).
Bornstein, P. Diversity of function is inherent in matricellular proteins: an appraisal of thrombospondin 1. J. Cell. Biol. 130, 503–506 (1995).
Yang, Z., Kyriakides, T. R. & Bornstein, P. Matricellular proteins as modulators of cell-matrix interactions: adhesive defect in thrombospondin 2-null fibroblasts is a consequence of increased levels of matrix metalloproteinase-2. Mol. Biol. Cell. 11, 3353–3364 (2000).
Noji, Y., Kajinami, K., Kawashiri, M. A., Todo, Y., Horita, T., Nohara, A. et al. Circulating matrix metalloproteinases and their inhibitors in premature coronary atherosclerosis. Clin. Chem. Lab. Med. 39, 380–384 (2001).
Kyriakides, T. R., Zhu, Y.- H., Smith, L. T., Bain, S. D., Yang, Z., Lin, M. T. et al. Mice that lack thrombospondin 2 display connective tissue abnormalities that are associated with disordered collagen fibrillogenesis, an increased vascular density, and a bleeding diathesis. J. Cell Biol. 140, 419–430 (1998).
Acknowledgements
We are most grateful to all the individuals who participated in this study and the collaborating clinics and physicians for referring individuals to the study. We thank Professor Momiao Xiong of Human Genetics Center, The University of Texas Health Science Center (Houston, TX, USA) for his encouraging discussion, Professor Aijun Sun of the Department of Cardiology, Zhongshan Hospital, Fudan University for their cooperation in collecting blood samples. The work was supported by the grants from Chinese High-Tech Program (2006AA020706, 2006AA02A406), Chinese National Natural Science Fund for Distinguished Young Scholars (30625019) and Shanghai Science and Technology Committee (09DZ2291900).
Author information
Authors and Affiliations
Corresponding author
Additional information
Supplementary Information accompanies the paper on Journal of Human Genetics website
Supplementary information
Rights and permissions
About this article
Cite this article
Wang, Y., Fu, W., Xie, F. et al. Common polymorphisms in ITGA2, PON1 and THBS2 are associated with coronary atherosclerosis in a candidate gene association study of the Chinese Han population. J Hum Genet 55, 490–494 (2010). https://doi.org/10.1038/jhg.2010.53
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/jhg.2010.53
Keywords
This article is cited by
-
An Myh11 single lysine deletion causes aortic dissection by reducing aortic structural integrity and contractility
Scientific Reports (2022)
-
Investigation of candidate genes and mechanisms underlying obesity associated type 2 diabetes mellitus using bioinformatics analysis and screening of small drug molecules
BMC Endocrine Disorders (2021)
-
Association of circulating angiogenesis inhibitors and asymmetric dimethyl arginine with coronary plaque burden
Fibrogenesis & Tissue Repair (2015)
-
MiR-1246 promotes SiHa cervical cancer cell proliferation, invasion, and migration through suppression of its target gene thrombospondin 2
Archives of Gynecology and Obstetrics (2014)
-
Association of PON1 genotype and haplotype with susceptibility to coronary artery disease and clinical outcomes in dual antiplatelet-treated Han Chinese patients
European Journal of Clinical Pharmacology (2013)