Abstract
Meniere's disease (MD) is a disorder of the inner ear characterized by episodes of vertigo, tinnitus and fluctuating sensorineural hearing loss. Most MD cases are sporadic, but 5–15% of patients are familial following an autosomal dominant mode of inheritance with incomplete penetrance. We have previously identified a candidate gene region for MD on chromosome 12p12.3 using linkage analysis. We genotyped 15 Swedish families segregating familial MD (FMD) to further clarify the role of chromosome 12p in a larger cohort of families. Highly polymorphic marker loci were analyzed over the 16-Mb candidate region in affected and healthy family members as well as in control subjects. The results revealed allelic association between FMD and several individual polymorphic marker alleles and single-nucleotide polymorphisms. Moreover, a common three-marker haplotype spanning 1.48 Mb co-segregates with FMD in 60% of the families investigated, forming the core of a possible ancestral haplotype associated with FMD in Sweden.
Similar content being viewed by others
Introduction
Meniere's disease (MD) is a disorder of the inner ear characterized by recurrent spontaneous and episodic vertigo, with fluctuating sensorineural hearing loss, aural fullness and tinnitus. The cause is not known, but it is believed that endolymphatic hydrops is a pathophysiological condition closely associated with the disease.1, 2 The prevalence of MD range between 17 and 218 per 100 0003, 4, 5, 6 and an incidence of 47 per 100 000 was reported in a Swedish study.7 Although the majority of cases with MD are sporadic, various studies estimate the frequency of familial cases to be between 5 and 15%.8, 9, 10, 11, 12, 13 The inheritance pattern from the analysis of familial cases is likely to be autosomal dominant with a reduced penetrance. Anticipation including both earlier onset and tendency to more severe symptoms in successive generations has also been suggested from a few studies.10, 11, 14
Genetic factors in MD are now widely recognized, but no causative gene variants have been reported despite several suggested candidate genes. Early attempts to identify genetic factors associated with MD include the analysis of the histocompatibility antigens, but no consistent association has been verified.11, 15 Human non-syndromic deafness with vestibular dysfunction (DFNA9) shares several clinical symptoms with MD.16, 17, 18 DFNA9 segregates COCH gene mutations; however, no mutations in COCH have yet been identified in MD patients.19, 20 Other candidates that have been ruled out include the genes encoding Antiquitin (ATQ1) and Aquaporin 2 (AQP2), which are important for fluid balance and endolymph homeostasis.21, 22 A mutation in the α-adductin gene has been proposed as a possible mechanism leading to endolymphatic hydrops in MD.23 An association study has shown functional polymorphisms in the lymphoid protein phosphatase PTPN22 to be associated with bilateral MD.24 Another case–control study has suggested single-nucleotide polymorphism (SNP) variants in the KCNE potassium channel genes (KCNE1 and KCNE2) to be associated with increased susceptibility to MD.25 This has not been confirmed in an independent investigation that instead showed association between MD and several SNPs within the host cell factor C1 (HCFC1) gene.26 A recent publication also failed to find the association between polymorphisms in KCNE1 or KCNE3 and MD in a Caucasian population.27
We have previously identified a candidate region for familial MD (FMD) on chromosome 12p12.3 from linkage analysis using three large Swedish families segregating the disease.28 Furthermore, affected individuals in two of the families share a single haplotype spanning the linked region, suggesting a possible ancestral mutation. To further investigate the chromosome 12p12 region in FMD, we performed a genetic analysis in 15 multiplex families segregating MD in at least two individuals. We performed allelic association and haplotype analysis using microsatellite markers spanning 16 Mb on chromosome 12p. Our results support an allelic association between FMD and several marker alleles and SNPs, as well as a specific chromosome 12p haplotype.
Materials and methods
Subjects
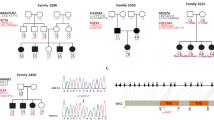
Blood samples were collected from 15 families with two or more affected members (including a subset of three families that have been reported with linkage to 12p12.3 in our previous study).28 Altogether, 24 affected and 14 unaffected individuals from the families segregating FMD were genotyped. Furthermore, a control group consisting of 31 ethnically matched family trios without any history of MD or partial MD symptoms were genotyped. All MD families and controls originated from Sweden and none of the families were known to be related. All individuals were clinically examined by CF or UF, and MD was diagnosed based on the diagnostic criteria of the Committee on Hearing and Equilibrium of the American Academy of Otolaryngology–Head and Neck Surgery.29 Informed consent was obtained from all participants and the study was approved by the ethics committee at Uppsala University.
Mutation analyses
The candidate gene RERGL, situated between D12S1595 and D12S301, and the PIK3C2G gene, situated between GT14 and TG21AG12, were directly sequenced on genomic DNA from three affected probands sharing the associated 12p haplotype (MD24, F1V1, F2IV1) and four controls. PCR products of all exons, exon–intron junctions and untranslated regions were sequenced using dye-terminator chemistry and separated on an ABI 3700 (ABI, Foster City, CA, USA). A U2 small nuclear RNA reported (NCBI, Ensemble, USCS) to be located in the candidate region was also investigated by sequencing. Any sequence variation identified, either in coding regions or close to coding regions, were used for association studies. Possible splice site alteration were investigated with the Splice Site Prediction by Neural Network.30 The primer sequences and the conditions for PCR are available upon request.
Allelic association and haplotype analyses
DNA was extracted from peripheral blood using standard protocols. Genotyping was performed using fluorescently labelled microsatellite markers as described previously.31 Altogether, we genotyped 24 microsatellite markers spanning a 16-Mb region between marker loci D12S1695 and D12S1617. Seven of these markers are novel polymorphic di-, tri- and tetranucleotide repeats selected from NCBI, build 36.3 (primer sequences, allele distribution and PCR conditions available upon request). Primers were designed using Primer 3 software (Whitehead Institute for Biomedical Research, http://frodo.wi.mit.edu/primer3/). Haplotypes were generated using Cyrillic 2.1 program (Cherwell Scientific Publishing, Oxford, UK) and confirmed manually. Haplotypes were assigned as ‘affected’ or ‘unaffected’ according to the segregation with MD in each family. We generated 15 ‘affected’ haplotypes each representing one MD family and 18 non-transmitted ‘control’ haplotypes. Moreover, 31 additional ‘control’ haplotypes were generated from unrelated non-affected families. Association between MD and individual markers/haplotypes was determined using Fisher exact test (two-tailed) and Pexcess. Fisher exact test was calculated using a 2 × 2 table with alleles classified into two groups, one for the associated allele, or haplotype, and all others combined into a single group according to the transmission/disequilibrium test.32 Pexcess represents an estimate of the proportion of disease chromosomes carrying the associated allele and is calculated as Pexcess=(Pa−Pn)/(1−Pn) where Pa is the frequency of the associated allele on disease chromosomes and Pn is the frequency of the same allele on normal chromosomes.33 SNPs identified by sequencing showing variation in our patient or control material were investigated for association to MD. The patient group consisted of probands from each of the MD families (n=14). One proband from each of the unrelated non-affected families was used as controls (n=18). We also compared the SNP genotype and allele frequencies with the HapMap Caucasian European from Utah (CEU) database (Utah residents with Northern and Western European ancestry from the Centre d'Etude du Polymorphisme Humain (CEPH)collection). Genotype frequencies of SNPs were compared by 2 × 3 contingency tables (AA, AB and BB genotypes) and allele frequencies of SNPs were compared by 2 × 2 contingency tables (major allele A versus minor allele B) using Fisher exact test (two tailed).
Results
Mutation analysis
No sequence alteration was found in the RERGL gene or U2 small nuclear RNA in the three affected probands when compared with controls and public databases. No genetic variants showing variation in our patient material could be identified in the RERGL gene or U2 small nuclear RNA. Sequence analysis of the PIK3C2G gene identified 11 coding or non-coding SNPs, including two novel SNPs, c.*28A>del and c.*56G>A, in the 3′ untranslated region, showing variance in our patient/control material. The novel c.*28A>del could possibly be the same as the reported SNP rs16914368 annotated as c.*28A>T. As the following base, c.*29, is a T, some sequencing methods would read c.*28A>del as c.*28A>T.
Allelic association and haplotype analysis
The 15 families were investigated for the 11 markers included in the haplotype previously reported segregating with MD in two families.28 To refine the region, we genotyped an additional 13 markers including six markers selected from public databases (NCBI, build 36.3) and seven novel polymorphic repeats. The previously reported shared haplotype in two of our families could also be confirmed and using the additional markers this shared haplotype now spans 1.6 Mb between marker D12S1630 and GT27. Allelic association and haplotype analysis were performed using in total 24 microsatellite marker alleles in order to investigate possible association with the 12p region in all MD families. Alleles from three individual marker loci, D12S363, D12S1595 and GATA20, showed significant allelic association to FMD ascertained by both Pexcess and Fisher exact test (Pexcess >0.4, P<0.05) to FMD (Table 1). Furthermore, we analyzed the association of all continuous three-marker haplotypes in the region. Five three-marker haplotypes were significantly associated with MD with the strongest association obtained for the D12S1595–D12S301–TA35 haplotype (Pexcess=0.464, P<0.01, Table 2). This three-marker haplotype was present in 53% of the affected chromosomes and on 13% of the control chromosomes. The haplotype is flanked by several three-marker haplotypes showing allelic association (Pexcess >0.4, P<0.05) of which the three-marker haplotype TA35-TA171-TC33 is the most prevalent in FMD families (60%). Expansion of the haplotypes identified two overlapping haplotypes spanning seven markers (D12S363 to TC33, Pexcess=0.357, P=0.011 and TA35 to GT11, Pexcess=0.333, P<0.01) each present in 40% of the MD families. Furthermore, 33% of the families, including the two families previously described,28 share a haplotype comprising 11 markers (D12S363 to GT11, Pexcess=0.379, P<0.01; Table 2). Considering both single marker and haplotype associations, the region spanning 1.48 Mb between the markers D12S373 and GT27 is strongly associated with MD in the Swedish population. Two genes are positioned within this region, the RERG/RAS-like gene (RERGL) and the PIK3C2G gene. Furthermore, we investigated the association of 11 SNPs (rs55845540, rs11044004, rs7133666, rs34734874, rs12309666, rs12827507, rs11044211, rs61757718, rs35889592, and the two novel SNPs c.*28A>del and c.*56G>A) within the PIK3C2G gene. SNP analysis identified two SNPs (rs12827507 and rs11044211) flanking exon 29 of PIK3C2G, showing both genotypic and allelic association to MD compared with both our control material and HapMap CEU (Supplementary Table 1). These SNPs are however not situated close enough to exon 29 to cause any splice site alterations (rs12827507 acceptor splice site score of 0.98, rs11044211 donor splice site score of 1.00).
Discussion
We present further evidence for a susceptibility locus behind FMD on chromosome 12p12. 3 using association analysis. Two out of three families in our previous study shared a single haplotype within the linked region and we investigated the region in a larger cohort of FMD using allelic association. Genotyping of individual markers revealed three alleles strongly associated with FMD, within the previously identified region on chromosome 12. Furthermore, several adjacent markers show relatively high degree of allelic association in the region. Haplotype analysis identified a region of 1.48 Mb between the markers D12S373 and GT27 on chromosome 12 strongly associated with FMD. In conclusion, our results confirm the previously reported susceptibility locus for FMD on chromosome 12 and suggest an ancestral locus for FMD in the Swedish population.
Two genes are positioned in the refined susceptibility locus, the RERG/RAS-like gene (RERGL), and part of the previously sequenced PIK3C2G gene.28 RERGL binds to GDP/GTP and may possess intrinsic GTPase activity, making it another likely candidate for MD.34 However, none of these genes show any sequence alterations in the coding sequences associated to MD. Non-coding or non-genic sequence alterations within the candidate region could not be ruled out. In fact two non-coding SNPs, rs12827507 (c.3889-41A>G) and rs11044211 (c.3964+25C>T), situated on either side of exon 29 of the PIK3C2G gene showed strong association to MD in our material. These were the only SNPs where the minor allele is associated to the MD haplotype and whether these confer to disease susceptibility remain unclear.
No gene causing non-syndromic hearing loss on 12p has yet been identified. However, in 2006, non-syndromic recessive hearing loss, DFNB62, was mapped to 12p13.2-11.23 in a Pakistani family.35 This region includes the region identified in the Swedish FMD families. The audiophenotype of DFNB62 was congenital profound hearing loss, but no vestibular dysfunction was described, making the probability that the genes are identical less likely. The failure to identify genes responsible for MD may suggest that the disease is a complex genetic condition with a contribution from coincidental gene variants. Alternatively, MD might derive from regulatory gene variants, which could explain the difficulty in identifying the underlying mutation(s) simply by examining coding or even transcribed sequences. Recent studies in mice and humans have established the importance of microRNAs (miRNAs) in the development and function of the inner ear.36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46 Point mutations in the seed region of miR-96, an miRNA expressed in hair cells of the inner ear, result in autosomal dominant, progressive hearing loss.43, 47 Mice mutant for several of the target genes for miRNAs exhibit deafness and hair cell degeneration, indicating the importance of this novel gene regulation in hearing loss.48, 49 To date, no intragenic miRNAs are annotated in the region associated with MD in our families. Further analysis is now required to clarify the molecular basis of the chromosome 12p12.3 region in MD, possibly with an emphasis on non-coding and non-genic sequences such as the two associated SNPs reported herein.
References
Yamakawa, K. Pathologic changes in a Ménière's patient. Proc. 42nd Meet. Jpn. Otorhinolaryngol. Soc. 44, 2310–2312 (1938).
Hallpike, C. Observations on the pathology of Meniere's disease. J. Laryngol. Otol. 53, 625–654 (1938).
Wladislavosky-Waserman, P., Facerm, G. W., Mokri, B. & Kurland, L. T. Meniere's disease: a 30-year epidemiologic and clinical study in Rochester, Mn, 1951-1980. Laryngoscope 94, 1098–1102 (1984).
Watanabe, Y., Mizukoshi, K., Shojaku, H., Watanabe, I., Hinoki, M. & Kitahara, M. Epidemiological and clinical characteristics of Meniere's disease in Japan. Acta Otolaryngol. 519, 206–210 (1995).
Celestino, D. & Ralli, G. Incidence of Meniere's disease in Italy. Am. J. Otol. 12, 135–138 (1991).
Cawthorne, T. & Hewlett, A. B. Meniere's disease. Proc. R. Soc. Med. 47, 663–670 (1954).
Stahle, J., Stahle, C. & Arenberg, I. K. Incidence of Meniere's disease. Arch. Otolaryngol. 104, 99–102 (1978).
Birgerson, L., Gustavson, K. H. & Stahle, J. Familial Meniere's disease: a genetic investigation. Am. J. Otol. 8, 323–326 (1987).
Martini, A. Hereditary Meniere's disease: report of two families. Am. J. Otolaryngol. 3, 163–167 (1982).
Morrison, A. W. Anticipation in Meniere's disease. J. Laryngol. Otol. 109, 499–502 (1995).
Arweiler, D. J., Jahnke, K. & Grosse-Wilde, H. [Meniere disease as an autosome dominant hereditary disease]. Laryngorhinootologie 74, 512–515 (1995).
Morrison, A. W. Meniere's disease. J. R. Soc. Med. 74, 183–189 (1981).
Klockars, T. & Kentala, E. Inheritance of Meniere's disease in the Finnish population. Arch. Otolaryngol. Head Neck Surg. 133, 73–77 (2007).
Fung, K., Xie, Y., Hall, S. F., Lillicrap, D. P. & Taylor, S. A. Genetic basis of familial Meniere's disease. J. Otolaryngol. 31, 1–4 (2002).
Koyama, S., Mitsuishi, Y., Bibee, K., Watanabe, I. & Terasaki, P. I. HLA associations with Meniere's disease. Acta Otolaryngol. 113, 575–578 (1993).
Robertson, N. G., Lu, L., Heller, S., Merchant, S. N., Eavey, R. D., McKenna, M. et al. Mutations in a novel cochlear gene cause DFNA9, a human nonsyndromic deafness with vestibular dysfunction. Nat. Genet. 20, 299–303 (1998).
Fransen, E., Verstreken, M., Verhagen, W. I., Wuyts, F. L., Huygen, P. L., D’Haese, P. et al. High prevalence of symptoms of Meniere's disease in three families with a mutation in the COCH gene. Hum. Mol. Genet. 8, 1425–1429 (1999).
Verstreken, M., Declau, F., Wuyts, F. L., D’Haese, P., Van Camp, G., Fransen, E. et al. Hereditary otovestibular dysfunction and Meniere's disease in a large Belgian family is caused by a missense mutation in the COCH gene. Otol. Neurotol. 22, 874–881 (2001).
Usami, S., Takahashi, K., Yuge, I., Ohtsuka, A., Namba, A., Abe, S. et al. Mutations in the COCH gene are a frequent cause of autosomal dominant progressive cochleo-vestibular dysfunction, but not of Meniere's disease. Eur. J. Hum. Genet. 11, 744–748 (2003).
Sanchez, E., Lopez-Escamez, J. A., Lopez-Nevot, M. A., Lopez-Nevot, A., Cortes, R. & Martin, J. Absence of COCH mutations in patients with Meniere disease. Eur. J. Hum. Genet. 12, 75–78 (2004).
Lynch, M., Cameron, T. L., Knight, M., Kwok, T. Y., Thomas, P., Forrest, S. M. et al. Structural and mutational analysis of antiquitin as a candidate gene for Meniere disease. Am. J. Med. Genet. 110, 397–399 (2002).
Mhatre, A. N., Jero, J., Chiappini, I., Bolasco, G., Barbara, M. & Lalwani, A. K. Aquaporin-2 expression in the mammalian cochlea and investigation of its role in Meniere's disease. Hear. Res. 170, 59–69 (2002).
Teggi, R., Lanzani, C., Zagato, L., Delli Carpini, S., Manunta, P., Bianchi, G. et al. Gly460Trp alpha-adducin mutation as a possible mechanism leading to endolymphatic hydrops in Meniere's syndrome. Otol. Neurotol. 29, 824–828 (2008).
Lopez-Escamez, J. A., Saenz-Lopez, P., Acosta, L., Moreno, A., Gazquez, I., Perez-Garrigues, H. et al. Association of a functional polymorphism of PTPN22 encoding a lymphoid protein phosphatase in bilateral Meniere's disease. Laryngoscope 120, 103–107 (2010).
Doi, K., Sato, T., Kuramasu, T., Hibino, H., Kitahara, T., Horii, A. et al. Meniere's disease is associated with single nucleotide polymorphisms in the human potassium channel genes, KCNE1 and KCNE3. ORL J. Otorhinolaryngol. Relat. Spec. 67, 289–293 (2005).
Vrabec, J. T., Liu, L., Li, B. & Leal, S. M. Sequence variants in host cell factor C1 are associated with Meniere's disease. Otol. Neurotol. 29, 561–566 (2008).
Campbell, C. A., Della Santina, C. C., Meyer, N. C., Smith, N. B., Myrie, O. A., Stone, E. M. et al. Polymorphisms in KCNE1 or KCNE3 are not associated with Meniere disease in the Caucasian population. Am. J. Med. Genet. A 152A, 67–74 (2010).
Klar, J., Frykholm, C., Friberg, U. & Dahl, N. A. Meniere's disease gene linked to chromosome 12p12.3. Am. J. Med. Genet. B Neuropsychiatr. Genet. 141B, 463–467 (2006).
Monsell, M. E., Balkany, A. T., Gates, A. G., Goldenberg, . R., Meyerhoff, L. W. & House, W. J. Committee on Hearing and Equilibrium guidelines for the diagnosis and evaluation of therapy in Meniere's disease. American Academy of Otolaryngology-Head and Neck Foundation, Inc. Otolaryngol. Head Neck Surg. 113, 181–185 (1995).
Reese, M. G., Eeckman, F. H., Kulp, D. & Haussler, D. Improved splice site detection in Genie. J. Comput. Biol. 4, 311–323 (1997).
Klar, J., Gedde-Dahl, JrT., Larsson, M., Pigg, M., Carlsson, B., Tentler, D. et al. Assignment of the locus for ichthyosis prematurity syndrome to chromosome 9q33.3-34.13. J. Med. Genet. 41, 208–212 (2004).
Spielman, R. S., McGinnis, R. E. & Ewens, W. J. Transmission test for linkage disequilibrium: the insulin gene region and insulin-dependent diabetes mellitus (IDDM). Am. J. Hum. Genet. 52, 506–516 (1993).
Hastbacka, J., de la Chapelle, A., Mahtani, M. M., Clines, G., Reeve-Daly, M. P., Daly, M. et al. The diastrophic dysplasia gene encodes a novel sulfate transporter: positional cloning by fine-structure linkage disequilibrium mapping. Cell 78, 1073–1087 (1994).
Kalinec, F., Zhang, M., Urrutia, R. & Kalinec, G. Rho GTPases mediate the regulation of cochlear outer hair cell motility by acetylcholine. J. Biol. Chem. 275, 28000–28005 (2000).
Ali, G., Santos, R. L., John, P., Wambangco, M. A., Lee, K., Ahmad, W. et al. The mapping of DFNB62, a new locus for autosomal recessive non-syndromic hearing impairment, to chromosome 12p13.2-p11.23. Clin. Genet. 69, 429–433 (2006).
Weston, M. D. & Soukup, G. A. MicroRNAs sound off. Genome Med. 1, 59 (2009).
Weston, M. D., Pierce, M. L., Rocha-Sanchez, S., Beisel, K. W. & Soukup, G. A. MicroRNA gene expression in the mouse inner ear. Brain Res. 1111, 95–104 (2006).
Tsonis, P. A., Call, M. K., Grogg, M. W., Sartor, M. A., Taylor, R. R., Forge, A. et al. MicroRNAs and regeneration: Let-7 members as potential regulators of dedifferentiation in lens and inner ear hair cell regeneration of the adult newt. Biochem. Biophys. Res. Commun. 362, 940–945 (2007).
Swiderski, D. L., Gong, T. W. & Mustapha, M. Complexity and integration in the control of inner-ear development. Genome Biol. 8, 315 (2007).
Soukup, G. A., Fritzsch, B., Pierce, M. L., Weston, M. D., Jahan, I., McManus, M. T. et al. Residual microRNA expression dictates the extent of inner ear development in conditional Dicer knockout mice. Dev. Biol. 328, 328–341 (2009).
Soukup, G. A. Little but loud: small RNAs have a resounding affect on ear development. Brain Res. 1277, 104–114 (2009).
Sacheli, R., Nguyen, L., Borgs, L., Vandenbosch, R., Bodson, M., Lefebvre, P. et al. Expression patterns of miR-96, miR-182 and miR-183 in the development inner ear. Gene Expr. Patterns 9, 364–370 (2009).
Mencia, A., Modamio-Hoybjor, S., Redshaw, N., Morin, M., Mayo-Merino, F., Olavarrieta, L. et al. Mutations in the seed region of human miR-96 are responsible for nonsyndromic progressive hearing loss. Nat. Genet. 41, 609–613 (2009).
Fritzsch, B., Beisel, K. W., Pauley, S. & Soukup, G. Molecular evolution of the vertebrate mechanosensory cell and ear. Int. J. Dev. Biol. 51, 663–678 (2007).
Friedman, L. M., Dror, A. A., Mor, E., Tenne, T., Toren, G., Satoh, T. et al. MicroRNAs are essential for development and function of inner ear hair cells in vertebrates. Proc. Natl Acad. Sci. USA 106, 7915–7920 (2009).
Friedman, L. M. & Avraham, K. B. MicroRNAs and epigenetic regulation in the mammalian inner ear: implications for deafness. Mamm. Genome 20, 581–603 (2009).
Lewis, M. A., Quint, E., Glazier, A. M., Fuchs, H., De Angelis, M. H., Langford, C. et al. An ENU-induced mutation of miR-96 associated with progressive hearing loss in mice. Nat. Genet. 41, 614–618 (2009).
Wallis, D., Hamblen, M., Zhou, Y., Venken, K. J., Schumacher, A., Grimes, H. L. et al. The zinc finger transcription factor Gfi1, implicated in lymphomagenesis, is required for inner ear hair cell differentiation and survival. Development 130, 221–232 (2003).
Goodyear, R. J., Legan, P. K., Wright, M. B., Marcotti, W., Oganesian, A., Coats, S. A. et al. A receptor-like inositol lipid phosphatase is required for the maturation of developing cochlear hair bundles. J. Neurosci. 23, 9208–9219 (2003).
Author information
Authors and Affiliations
Corresponding author
Additional information
Supplementary Information accompanies the paper on Journal of Human Genetics website
Supplementary information
Rights and permissions
About this article
Cite this article
Gabriková, D., Frykholm, C., Friberg, U. et al. Familiar Meniere's disease restricted to 1.48 Mb on chromosome 12p12.3 by allelic and haplotype association. J Hum Genet 55, 834–837 (2010). https://doi.org/10.1038/jhg.2010.122
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/jhg.2010.122
Keywords
This article is cited by
-
Genetic advances in Meniere Disease
Molecular Biology Reports (2023)
-
Variable expressivity and genetic heterogeneity involving DPT and SEMA3D genes in autosomal dominant familial Meniere’s disease
European Journal of Human Genetics (2017)
-
M. Menière: Diagnostische Kriterien des Internationalen Klassifikationskomitees der Bárány-Gesellschaft
HNO (2017)