Abstract
Cleft of the soft palate (CSP) and the hard palate are subtypes of cleft palate. Patients with either condition often have difficulty with speech and swallowing. Nonsyndromic, cleft palate isolated has been reported to be associated with several genes, but to our knowledge, there have been no detailed genetic investigations of CSP. We performed a genome-wide linkage analysis using a single-nucleotide polymorphism-based microarray platform and successively using microsatellite markers in a family in which six members, across three successive generations, had CSP. A maximum LOD score of 2.408 was obtained at 2p24.2–24.1 and 2p21–p12, assuming autosomal dominant inheritance. Our results suggest that either of these regions is responsible for this type of CSP.
Similar content being viewed by others
Introduction
Orofacial cleft, one of the most common congenital malformations, is a heterogeneous group of complex traits. Orofacial cleft is classified into two main categories, cleft lip with or without cleft palate and cleft palate isolated (CPI). Both clefting phenotypes can appear to be related to some syndromes (syndromic orofacial cleft) or not be related to syndromes (nonsyndromic orofacial cleft). CPI is considered genetically distinct from cleft lip with or without cleft palate, on the basis of epidemiological evidence and the different developmental timing of lip and palate formation. Recent molecular genetic studies1, 2, 3, 4, 5 have identified genes or loci that are responsible for CPI. However, fewer genes and/or loci-associated CPI have been reported in comparison with cleft lip with or without cleft palate.6
CPI is mostly classified into two subtypes morphologically: cleft of the hard palate (CHP) and cleft of the soft palate (CSP).7 Submucous cleft palate (SMCP) is a small subgroup in the CPI. SMCP manifests with bifid uvula, separation of the muscle with an intact mucosa and a bony defect in the posterior edge of the hard palate.8 Both CHP and CSP are caused by a failure of fusion of the palatal shelves, but little is known about the cause of the difference in their phenotypes. Christensen et al.9 suggested that CHP and CSP might be etiologically distinct. Although patients with CSP have serious problems in speech and deglutition, as well as CHP, there have been no detailed genetic studies performed.
We recently encountered a Japanese family that included five CSP patients and one SMCP patient. The aim of this study was to identify the CSP/SMCP predisposing locus in this family using genome-wide single-nucleotide polymorphism (SNP)-based linkage analysis.
Materials and methods
Family and patients
A Japanese family included five patients (I-2, II-2, II-3, III-1 and III-2) with CSP and one patient (II-5) with SMCP across three generations (Figure 1). Two patients (II-2 and II-3) were monozygotic twins. The phenotypes of two patients (III-1 and II-5) were shown in Figure 2. All patients had no other symptoms such as mental retardation, and all family members were examined by one or two well-trained dentists.
Family tree with haplotypes at 2p24.2–24.1 (CSPR1) and 2p21–p12 (CSPR2). Black closed, gray closed and open symbols indicate affected with cleft of the soft palate (CSP), affected with subcutaneous cleft palate (SMCP) and unaffected, respectively. An arrow indicates the proband. Genotypes of microsatellite markers defining the candidate intervals are shown below each individual. Boxed haplotype indicates possibly disease-associated haplotype.
The disease in the family was consistent with an autosomal dominant mode of inheritance. Blood samples were obtained with written informed consent from 15 cooperative family members (Figure 1). The study protocol was approved by the committee for ethical issues on the Human Genome and Gene Analysis of Nagasaki University.
SNP genotyping and linkage analysis
Genomic DNA was extracted from peripheral blood lymphocytes of the 15 members, using a QIAamp DNA Mini Kit (QIAGEN, Hilden, Germany). Their genotypes were determined using a GeneChip Human Mapping 10K 2.0 Xba Array (Affymetrix, Santa Clara, CA, USA). We used MERLIN software10 to analyze compiled pedigree data sets. Mendelian errors were detected by PEDCHECK,11 and SNPs with Mendelian error were not used in the data analysis. LOD scores were calculated under a parametric autosomal dominant model in which penetrance was set to 1.0 and disease allele frequency was 0.00001. As CSP and SMCP can be categorized together because of their similar anatomical features,9 the patient with SMCP and the patients with CSP (II-5) were classified as ‘affected’ for linkage score calculations.
To confirm the result of linkage data using the GeneChip Human Mapping 10K 2.0 Xba Array, we performed a two-point linkage analysis using microsatellite markers by the method described elsewhere.12 The two-point LOD score was calculated using MLINK program.13
Results and discussion
In the assay with the 10K-Array, the GeneChip call rates varied from 92.18 to 99.42% (with a mean of 97.54%). Two regions, 2p24.2–p24.1 (CSP region 1: CSPR1), a 4.5-Mb interval between rs1545497 and rs1872325, and 2p21–p12 (CSP region 2: CSPR2), a 34.5-Mb segment between rs940053 and rs310777, were CSP candidate loci with a maximum LOD score of 2.408 (Figure 3). The LOD scores of all other regions were below 1.000. Two-point LOD scores using microsatellite markers showed the same scores (2.408); therefore, the result of linkage analysis from SNP genotyping was reconfirmed (haplotype using microsatellite markers was shown in Figure 1). It is thus likely that a gene having a role in palatal fusion is located within either CSPR1 or CSPR2.
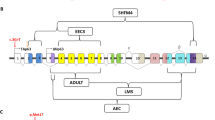
On the basis of our knowledge of oral palate development, we chose nine genes from the candidate CSP regions and performed mutation analysis. Of the nine candidate genes, three were from CSPR1: growth/differentiation factor 7 (GDF7), matrilin 3 (MATN3) and member B of the Ras homolog gene family (RHOB). The other six genes were from CSPR2: calmodulin 2 (CALM2), bone morphologic protein 10 (BMP10), sprouty-related EVH1 domain-containing protein 2 (SPRED2), transforming growth factor, alfa (TGFA), ventral anterior homeobox 2 (VAX2; 2p13.3) and stoned B-like factor/stonin 1 (STON1). Most of these genes are concerned with bone development, and with the transforming growth factor and mitogen-activated protein kinase signaling pathways, or are transcription factors related to homeobox genes. However, no pathogenic mutation was found within any of its exons or intron–exon boundaries of all nine genes.
To detect structural genomic alterations that may cause CSP within the candidate regions, we performed copy number analysis with the proband's DNA using the Genome-Wide Human SNP Array 5.0 (Affymetrix). Although several copy number alterations were detected (data were not shown), all were already registered as copy number variations on the UCSC Genome Browser (http://genome.ucsc.edu/) and none of them coincided with regions with positive LOD scores.
In conclusion, this is the first report of a whole-genome linkage analysis scan for CSP. Although the LOD scores calculated are not high enough to assign the disease locus definitively, our data suggest that it lies at either 2p24.2–24.1 or 2p21–p12.
References
FitzPatrick, D. R., Carr, I. M., McLaren, L., Leek, J. P., Wightman, P., Williamson, K. et al. Identification of SATB2 as the cleft palate gene on 2q32-q33. Hum. Mol. Genet. 12, 2491–2501 (2003).
Braybrook, C., Doudney, K., Marçano, A. C., Arnason, A., Bjornsson, A., Patton, M. A. et al. The T-box transcription factor gene TBX22 is mutated in X-linked cleft palate and ankyloglossia. Nat. Genet. 29, 179–183 (2001).
Koillinen, H., Lahermo, P., Rautio, J., Hukki, J., Peyrard-Janvid, M. & Kere, J. A. Genome-wide scan of non-syndromic cleft palate only (CPO) in Finnish multiplex families. J. Med. Genet. 42, 177–184 (2005).
Shiang, R., Lidral, A. C., Ardinger, H. H., Buetow, K. H., Romitti, P. A. & Munger, R. G. Association of transforming growth-factor alpha gene polymorphisms with nonsyndromic cleft palate only (CPO). Am. J. Hum. Genet. 53, 836–843 (1993).
Hwang, S. J., Beaty, T. H., Panny, S. R., Street, N. A., Joseph, J. M., Gordon, S. et al. Association study of transforming growth factor alpha (TGF alpha) TaqI polymorphism and oral clefts: indication of gene-environment interaction in a population-based sample of infants with birth defects. Am. J. Epidemiol. 141, 629–636 (1995).
Carinci, F., Scapoli, L., Palmieri, A., Zollino, I. & Pezzetti, F. Human genetic factors in nonsyndromic cleft lip and palate: An update. Int. J. Pediatr. Otorhinolaryngol. 71, 1509–1519 (2007).
Millard, D. R. in Cleft Craft, the Evolution of its Surgery, Vol 1 The Unilateral Deformity (eds Millard, D.R.) 41–55 (Little Brown, Boston, 1976).
Calnan, J. Submucous cleft palate. Br. J. Plast. Surg. 6, 264–282 (1954).
Christensen, K. & Fogh-Anderson, P. Etiological subgroups in non-syndromic isolated cleft palate. A genetic-epidemiological study of 52 Danish birth cohorts. Clin. Genet. 46, 329–335 (1994).
Abecasis, G. R., Cherny, S. S., Cookson, W. O. & Cardon, L. R. Merlin--rapid analysis of dense genetic maps using sparse gene flow trees. Nat. Genet. 30, 97–101 (2002).
O’Connell, J. R. & Weeks, D. E. PedCheck: a program for identification of genotype incompatibilities in linkage analysis. Am. J. Hum. Genet. 63, 259–266 (1998).
Nakashima, M., Nakano, M., Hirano, A., Kishino, T., Kondoh, S., Miwa, N. et al. Genome-wide linkage analysis and mutation analysis of hereditary congenital blepharoptosis in a Japanese family. J. Hum. Genet. 53, 34–41 (2008).
Lathrop, G. M., Lalouel, J. M., Julier, C. & Ott, J. Strategies for multilocus linkage analysis in humans. Proc. Natl Acad. Sci. USA 81, 3443–3446 (1984).
Acknowledgements
We are grateful to the members of the family for their participation in this research. We also thank Ms Miho Ooga and Ms Chisa Hayashida for their technical assistance. KY was supported partly by a Grant-in-Aid for Scientific Research from the Ministry of Health, Labour and Welfare, and partly by grants from the Takeda Scientific Foundation and the Naito Foundation.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Tsuda, M., Yamada, T., Mikoya, T. et al. A type of familial cleft of the soft palate maps to 2p24.2–p24.1 or 2p21–p12. J Hum Genet 55, 124–126 (2010). https://doi.org/10.1038/jhg.2009.131
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/jhg.2009.131