Abstract
L-2-hydroxyglutaric aciduria (L-2-HGA, MIM 236792) is a neurometabolic disorder caused by the toxic accumulation of high concentration of L-2-hydroxyglutaric acid in plasma and cerebrospinal fluid. Distinct mutations on the L2HGDH gene have been associated with the clinical and biochemical phenotype. Here we present three novel mutations (Gln197X, Gly211Val and c.540+1 G>A), which increase the present deleterious collection of L2HGDH gene up to 35 mutations that we have compiled in this study. In addition, we used the haplotypic information based on polymorphic markers to demonstrate the common origin of Gly57Arg harboring chromosomes.
Similar content being viewed by others
Main
L-2–hydroxyglutaric aciduria (L-2-HGA; MIM 236792) is a rare neurometabolic disorder biochemically characterized by the presence of high levels of L-2-hydroxyglutaric acid in plasma, cerebrospinal fluid and urine.1 Although the biochemical hallmark, the L-2-hydroxyglutaric acid, was identified approximately three decades ago, the identification of both molecular and enzymatic defects was only recently established. It is now known that the mutated gene in L-2-HGA affects subject maps at chromosome 14q222, 3 and encodes a FAD-dependent enzyme2 known as L-2-hydroxyglutaric acid dehydrogenase.2, 4, 5
Once clarified the molecular basis involved in L-2-HGA, the reports of co-segregating mutations started to increase and are now available for several populations. At the time of this study, 32 distinct mutations found in a total of 50 affected families are known from several distinct populations.2, 3, 6, 7, 8, 9, 10 An interesting outcome of these population-specific screenings is that, in addition to the wide mutational spectrum of L2HGDH gene, high levels of population heterogeneity concerning the type and frequency of each mutation are also observed. To the best of our knowledge, only Pro302Leu has been repeatedly documented in different populations3, 6, 7 remaining unknown if the affected chromosomes share a common origin, or alternatively, if it has occurred through distinct mutational events.
As pathological heterogeneity exists when distinct populations are considered, a better and refined understanding of the mutational dynamics at L2HGDH locus would welcome more population-based studies. In this perspective, we investigated six previously unreported patients belonging to affected families of Portuguese (families 1 and 2), Brazilian (family 3) and Italian (families 4 and 5) ancestry following routinely molecular procedures, as previously described.6 Informed consent was obtained for all the collections. All the patients showed high levels of L-2-hydroxyglutaric acid in urine. Clinical phenotype was documented as follows:
Family 1: The patient is the second child of healthy non-consanguineous Portuguese individuals. A tonic–clonic seizure was noticed within 23 months of age and not repeated. The EEG was normal. Relative macrocephaly and slight global psychomotor retardation were recorded. By 4 years of age, developmental coefficient was 80. Magnetic resonance imaging (MRI) of the brain at 4 years of age disclosed subcortical leukodystrophy and hyperintensity of the cerebellar nuclei. At present, except for moderate mental retardation and macrocephaly, physical examination is normal.
Family 2: The patient is the second child of healthy non-consanguineous Portuguese couple. Slight psychomotor retardation with independent walking by 18–24 months and speaking by 18–36 months of age and slight kinetic tremor were reported. By 4 years of age gait disturbance caused orthopedic consultation. She was first observed at 6 years because of ataxia, intentional tremor, dysmetry, disartry and poor speech. At 8 years, the cerebral CT showed leukodystrophy. The brain MRI at 10 years of age showed subcortical leukodystrophia. At 10 and 12 years of age, global IQ was 69 and 62, respectively. Progressive gait disturbance, with pyramidal syndrome ensued.
Family 3: The two sister patients, currently at 30 and 33 years of age, were born from consanguineous Brazilian parents. Both present similar clinical phenotype. Specifically, both have cognitive retardation, epilepsy and axial ataxia. The brain MRI showed leukodystrophy suggestive of L-2-HGA.
Family 4: The patient was born at term after an uneventful pregnancy. Consanguineous parents (second cousins) are healthy individuals. In the first few months after birth a psychomotor delay was evident. MRI at 4 years of age revealed generalized white-matter signal abnormalities in the cerebral subcortical regions (imaging features of a subcortical leukoencephalopathy) with a loss of signal on T1-weighted slices and an increase in signal intensity on T2-weighted images. The corpus callosum appeared normal. The dentate nuclei showed abnormally high T2-weighted signals; also cerebellar vermis hypoplasia was observed. At present, the patient's clinical picture is being characterized by psychomotor delay (IQ=47), intentional tremor, motor dysfunction with ataxia, pyramidal and extrapyramidal signs. He is able to walk without support.
Family 5: The patient is the only child of healthy unrelated parents of Italian ancestry. No significant abnormalities were noted in the first year of life, but a slight developmental delay became evident after the first year. At the age of 5 years, he experienced a febrile generalized seizure. The MRI showed white-matter signal abnormalities in cerebral subcortical regions (subcortical leukoencephalopathy) with the involvement of globus pallidus without thalamic lesion. The periventricular white matter was less involved, and the corpus callosum appeared normal on T2-weighted sections. A hyperintense signal in the anterior limbs of the internal capsules and the entire external capsules was noted. Diffuse abnormalities in the cerebellum (dentate nuclei) were observed. Slight atrophy cortical was present. At 7 years of age, because of recurrent epileptic episodes, therapy with anti-epileptic treatment (valproic acid and clobazam) was started with good response. At present, the patient is showing mild psychomotor delay (IQ=63), tremor, mild motor dysfunction with ataxia. He is able to walk without support.
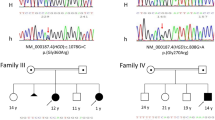
Once the clinical and biochemical phenotype had been established, molecular studies were performed in an attempt to detect the disease-associated mutation. The resulting data are presented in Table 1. A total of five distinct mutations were identified, three of which represented novel disease-associated mutations (Gln197Stop, Gly211Val and c.540+1 G>A), and the remaining (His98Arg and Gly57Arg) corresponding to previously documented replacements.6 Regarding the Gly211Val, the replacement of a small conserved residue by a long side-chain valine might have some impact on the structure of the protein. No studies were possible in the Brazilian control population, although both adult sisters show phenotypic features of L-2-HGA.
In what concerns the c.540+1 G>A and Gln197Stop replacements found in the same patient (family 5), though it is impossible to predict the impairment that a mutation affecting the consensus GT donor splice site11 would have because alternative spliced forms may be formed, we foresee a complete absence of activity caused by the p.Gln197Stop mutation which would lead to a truncated non-functional transcript. It should also be mentioned that the screening of 102 Portuguese unrelated health individuals did not reveal a single case harboring any of these mutations.
By adding these three new mutations to the currently known mutational repertoire of L2HGDH, the number of disease-associated replacements reaches a total of 35 (Table 2), and 18 of them are listed at the Human Gene Mutation Database (HGMD).12 The statistical information when all deleterious mutations are taken into account can be summarized as follows: 43% of all mutations (15 out of 35) are amino-acid replacements, 20% (7 out of 35) originate premature stop codons, about 17% (6 out of 35) represent small insertions or deletions (indels), 14% (5 out of 35) replace intronic splicing sequences, and just 6% (2 out of 35) are large deletions but technical difficulties in their detection may be covering a higher fraction.
In the absence of crystal structures for human or other mammalian species, we attempted to infer the relative impact that non-synonymous replacements may have by using a comparative protein-sequence approach. In order to accomplish this, homologous sequences representative of eukaryotic and prokaryotic species were extracted from KEGG (Kyoto Encyclopedia of Genes and Genomes13, 14, 15) or NCBI (National Center for Biotechnology Information at http://www.ncbi.nlm.nih.gov) databases and aligned using Geneious v4.016 (Figure 1). We observed that the 14 disease-associated residues, including the newly Gly211Val, lie in strongly conserved positions. In fact, nine mutations (Gly55Asp, Gly57Arg, Lys81Glu, His98Tyr, Glu176Asp, Tyr195Cys, Gly211Val, Pro302Leu and Ser440Tyr) replace invariant sites in both eukaryotes and prokaryotes and the remaining five mutations (Ala62Asp, Gly156Val, Ser263Leu, Val296Glu and His434Pro) lie in conserved sites across all eukaryotic species. This observation points to the relevance of these residues to the function of the protein at the same time that raises the possibility that replacements affecting less conserved positions are present in the general population, as they would result in mildly, possibly misidentified, clinical phenotypes.
Conservation profile of L2HGDH sequences across eukaryote and prokaryote species. Residues associated with disease-causing replacements are highlighted. Protein sequences were extracted from KEGG (Homo sapiens 79944, Canis familiaris 480316 and Gallus gallus 423573). The remaining protein sequences were obtained from National Center for Biotechnology Information (NCBI) as follows: Bos taurus (AAI51578), Pongo abelii (NP_001125894), Rattus norvegicus (NP_001101498) and Mus musculus (NP_663418), Synechocystis sp. (NP_442886) and Escherichia coli (NP_289209).
As mentioned before, mutations shared by different populations are rare. Yet, we are presenting an Italian patient homozygous for the Gly57Arg mutation, which was previously found at heterozygosity state in a Portuguese female (patient 10 in Vilarinho et al.6). To distinguish between a common and an independent origin we have used the intragenic polymorphic information.6 This analysis revealed that both patients share the same haplotypic background in maternally and paternally inherited chromosomes resulting in homozygosity for all the eight single nucleotide polymorphic sites. More specifically, the genetic background correspond to the following genomic sequence coordinates: T53(exon 1)-C159(exon 2)-A408+36nt(intron 3)-A533(exon 4)-C541−28nt(intron 4)-T703+12nt(intron 5)-G1196+13nt(intron 9)-C1196+34(intron 9). This strongly suggests that Portuguese and Italian Gly57Arg mutation-carrying chromosomes share a common origin, although extension of the haplotype with additional polymorphic data could increase the degree of confidence in this hypothesis. For the moment, it is noteworthy that a rare mutation, such as Gly57Arg, has been passing through geographically distinct populations, whereas more common ones are still confined to specific population groups.
To conclude, in this report we detail the molecular diagnosis of six new L-2-HGA patients, three of them harboring novel disease-associated mutations (p.Gln197Stop, c.540+1 G>A and p.Gly211Val). To date, the mutational spectrum of the L2HGDH gene has revealed 35 distinct mutations and, hopefully, upcoming population surveys will complete this collection for a better understanding of this disease.
References
Duran, M., Kamerling, J., Bakker, H., van Gennip, A. & Wadman, S. L-2-Hydroxyglutaric aciduria: an inborn error of metabolism? J. Inherit. Metab. Dis. 3, 109–112 (1980).
Rzem, R., Veiga-da-Cunha, M., Noel, G., Goffette, S., Nassogne, M C., Tabarki, B. et al. A gene encoding a putative FAD-dependent l-2-hydroxyglutarate dehydrogenase is mutated in l-2-hydroxyglutaric aciduria. Proc. Natl. Acad. Sci. USA 101, 16849–16854 (2004).
Topçu, M., Jobard, F., Halliez, S., Coskun, T., Yalcinkayal, C., Gerceker, F. O. et al. L-2-Hydroxyglutaric aciduria: identification of a mutant gene C14orf160, localized on chromosome 14q22.1. Hum. Mol. Genet. 13, 2803–2811 (2004).
Rzem, R., Van Schaftingen, E. & Veiga-da-Cunha, M. The gene mutated in l-2-hydroxyglutaric aciduria encodes l-2-hydroxyglutarate dehydrogenase. Biochimie 88, 113–116 (2006).
Rzem, R., Vincent, M. F., Van Schaftingen, E. & Veiga-da-Cunha, M. L-2-Hydroxyglutaric aciduria, a defect of metabolite repair. J. Inherit. Metab. Dis. 30, 681–689 (2007).
Vilarinho, L., Cardoso, M. L., Gaspar, P., Barbot, C., Azevedo, L., Diogo, L. et al. Novel L2HGDH mutations in 21 patients with L-2-hydroxyglutaric aciduria of Portuguese origin. Hum. Mut. 26, 395–396 (2005).
Sass, J. O., Jobard, F., Topçu, M., Mahfoud, A., Werlé, E., Cure, S. et al. L-2-Hydroxyglutaric aciduria: identification of ten novel mutations in the L2HGDH gene. J. Inherit. Metab. Dis. 105 (2008).
Larnaout, A., Amouri, R., Kefi, M. & Hentati, F. L-2-Hydroxyglutaric aciduria: clinical and molecular study in three Tunisian families. Identification of a new mutation and inter-familial phenotype variability. J. Inherit. Metab. Dis. 4 (2008).
O’Connor, G., King, M., Salomons, G., Jakobs, C., King, M. & Salomons, G. A novel mutation as a cause of L-2-hydroxyglutaric aciduria. J. Neurol. 256, 672–673 (2009).
Haliloglu, G., Jobard, F., Oguz, K K., Anlar, B., Akalan, N., Coskun, T. et al. L-2-hydroxyglutaric aciduria and brain tumors in children with mutations in the L2HGDH gene: neuroimaging findings. Neuropediatrics 39, 119–122 (2008).
Shapiro, M. B. & Senapathy, P. RNA splice junctions of different classes of eukaryotes: sequence statistics and functional implications in gene expression. Nucl. Acids Res. 15, 7155–7174 (1987).
Stenson, P. D., Ball, E. V., Mort, M., Phillips, A. D., Shiel, J. A. & Thomas, N. S. T. The human gene mutation database (HGMD): 2003 update. Hum. Mutat. 21, 577–581 (2003).
Kanehisa, M. & Goto, S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucl. Acids Res. 28, 27–30 (2000).
Kanehisa, M., Araki, M., Goto, S., Hattori, M., Hirakawa, M., Itoh, M. et al. KEGG for linking genomes to life and the environment. Nucl. Acids Res. 36, D480–D484 (2008).
Kanehisa, M., Goto, S., Hattori, M., Aoki-Kinoshita, K. F., Itoh, M., Kawashima, S. et al. From genomics to chemical genomics: new developments in KEGG. Nucl. Acids Res. 34, D354–D357 (2006).
Drummond, A. J., Cheung, M., Heled, J., Kearse, M., Moir, R., Stones-Havas, S. et al. at http://www.geneious.com/ (2008).
Acknowledgements
LA (C2007-IPATIMUP/AA1) is supported by Fundação para a Ciência e a Tecnologia (FCT) Ciência 2007 and by European Social Fund. IPATIMUP is partially supported by ‘Programa Operacional Ciência e Inovação 2010’ (POCI 2010), VI Programa Quadro (2002–2006).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Vilarinho, L., Tafulo, S., Sibilio, M. et al. Identification of novel L2HGDH gene mutations and update of the pathological spectrum. J Hum Genet 55, 55–58 (2010). https://doi.org/10.1038/jhg.2009.110
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/jhg.2009.110
Keywords
This article is cited by
-
A novel protein truncating mutation in L2HGDH causes L-2-hydroxyglutaric aciduria in a consanguineous Pakistani family
Metabolic Brain Disease (2022)
-
Computational analysis of receptor tyrosine kinase inhibitors and cancer metabolism: implications for treatment and discovery of potential therapeutic signatures
BMC Cancer (2019)
-
Identification of novel L2HGDH mutation in a large consanguineous Pakistani family- a case report
BMC Medical Genetics (2018)
-
A novel compound heterozygous mutation of the L2HGDH gene in a Chinese boy with L-2-hydroxyglutaric aciduria: case report and literature review
Neurological Sciences (2018)