Abstract
H+/organic cation antiporters (multidrug and toxin extrusion: MATE1 and MATE2-K) play important roles in the renal tubular secretion of cationic drugs. We have recently identified a regulatory single nucleotide polymorphism (SNP) of the MATE1 gene (−32G>A). There is no other information about SNPs of the MATE gene. In this study, we evaluated the functional significance of genetic polymorphisms in MATE1 and MATE2-K. We sequenced all exons of MATE1 and MATE2-K genes in 89 Japanese subjects and identified coding SNPs (cSNPs) encoding MATE1 (V10L, G64D, A310V, D328A and N474S) and MATE2-K (K64N and G211V). All the variants except for MATE1 V10L showed significant decrease in transport activity. In particular, MATE1 G64D and MATE2-K G211V variants completely lost transport activities. When membrane expression level was evaluated by cell surface biotinylation, those of MATE1 (G64D and D328A) and MATE2-K (K64N and G211V) were significantly decreased compared with that of wild type. These findings suggested that the loss of transport activities of the MATE1 G64D and MATE2-K G211V variants were due to the alteration of protein expression in cell surface membranes. This is the first demonstration of functional impairment of the MATE family induced by cSNPs.
Similar content being viewed by others
Introduction
In the proximal tubules of the mammalian kidney, organic ion transporters limit or prevent the toxicity of organic anions and cations by actively secreting these substances from the circulation into the urine.1, 2, 3, 4 Among human organic ion transporters located at the basolateral membranes, organic cation transporter 2 (OCT2), organic anion transporter 1 (OAT1) and OAT3 were isolated a decade ago, and have been well characterized as key transporters to regulate the renal handling of ionic drugs.4, 5 In contrast, the molecular functions of apical transporters have been only recently characterized. For example, multidrug resistance-associated protein 4 (MRP4) was demonstrated to be responsible for the renal elimination of antiviral drugs,6 diuretics7 and cephalosporin antibiotics.8 Human orthologs of the multidrug and toxin extrusion (MATE) family, members of which confer multidrug resistance on bacteria, were identified most recently,9, 10 and named MATE1 (SLC47A1) and MATE2-K (SLC47A2). Both transporters are expressed mainly in the renal brush border membranes, and are able to transport tetraethylammonium (TEA) utilizing an oppositely directed H+ gradient as a driving force,11 indicating that MATE1 and MATE2-K are H+/organic cation antiporters. These findings have improved the molecular understanding of the transcellular transport of ionic drugs in the renal tubules.
It is widely recognized that there is a large variation in the responses to drugs among individuals. Many enzymes involved in drug metabolism, such as cytochrome P450 and uridine diphosphate-glucuronosyltransferase are known to be polymorphic and have been associated with variations in blood concentrations of drugs.12 In addition to drug-metabolizing enzymes, the clinical significance of genetic variation of drug transporters has been demonstrated.13 For example, polymorphisms of SLCO1B1, which encodes the organic anion transporting polypeptide 1B1 to mediate the hepatic uptake of pravastatin, contribute to the interindividual variability in the disposition of pravastatin.14 Recent studies of OCT have demonstrated that polymorphisms of the OCT1 gene in Caucasians and the renal OCT2 gene in Koreans are responsible for the interindividual differences in the therapeutic efficacy and pharmacokinetics of metformin, an anti-diabetic agent.15, 16, 17
Metformin showed large interindividual variation in renal clearance, and a potential genetic contribution by the renal transporter was speculated.18 Because metformin is also a superior substrate for MATE1 and MATE2-K,10, 19 polymorphisms of MATE1 and MATE2-K genes may be involved in the interindividual difference in the renal clearance. We have recently identified a single nucleotide polymorphism (SNP) in the promoter region of MATE1 (−32G>A), which causes a decrease in Sp1 binding and promoter activity of approximately 50%.20 However, other genetic information for these transporters, especially the polymorphisms in the coding region, and their effect on functional properties, have not been well evaluated. In this study, therefore, we screened for polymorphisms in all exons of MATE1 and MATE2-K genes, and examined their transport activities by in vitro transient expression system.
Materials and methods
Materials
[14C]TEA bromide (2.035 GBq mmol−1) and [14C]metformin (1.998 GBqmmol−1) were obtained from American Radiolabeled Chemicals Inc. (St Louis, MO, USA) and Moravek Biochemicals Inc. (Brea, CA, USA), respectively. All other chemicals used were of the highest purity available.
Identification of SNPs of MATE1 and MATE2-K genes
Genomic DNA was isolated from peripheral blood from 89 Japanese subjects with renal diseases using a Wizard Genomic DNA Purification Kit (Promega, Madison, WI, USA). Genotyping was investigated by direct sequencing. PCR primers were designed to span all 17 exons of MATE1 and MATE2-K (GenBank accession number NT_010718) (Table 1). The PCR conditions were 94 °C for 3 min, followed by 40 cycles of 94 °C for 30 s, 60 °C for 30 s and 72 °C for 30 s, and then a final extension at 72 °C for 10 min, except for MATE1 exon 1. The condition for MATE1 exon 1 was 94 °C for 1 min, followed by 35 cycles of 94 °C for 30 s and 68 °C for 3 min, and then a final extension at 68 °C for 3 min. The PCR products were sequenced using a multicapillary DNA sequencer RISA384 system (Shimadzu, Kyoto, Japan). This study was conducted in accordance with the Declaration of Helsinki and its amendments and was approved by the Ethics Committee of Kyoto University Graduate School and Faculty of Medicine. All subjects gave their written informed consents.
Construction of non-synonymous variants of MATE1 and MATE2-K
MATE1 and MATE2-K cDNA were excised from MATE1/pcDNA3.1 and MATE2-K/pcDNA3.1,10 and were subcloned into pcDNA3.1/nV5-DEST (Invitrogen, Carsbad, CA, USA) to yield nV5-MATE1 and nV5-MATE2-K. Non-synonymous variants were constructed by the site-directed mutagenesis of nV5-MATE1 and nV5-MATE2-K, using a QuikChange II Site-Directed Mutagenesis Kit (Stratagene, La Jolla, CA, USA) with the primers listed in Table 2. The nucleotide sequences of these constructs were confirmed using a multicapillary DNA sequencer RISA384 system (Shimadzu).
Transport studies
HEK293 cells (ATCC CRL-1573; American Type Culture Collection) were cultured in complete medium consisting of Dulbecco's modified Eagle's medium (Sigma Chemical Co., St Louis, MO, USA) with 10% fetal bovine serum (Invitrogen) in an atmosphere of 5% CO2 and 95% air at 37 °C. cDNA plasmid transfection (Figure 2: 25 ng; Figure 3: 100 ng; Figure 6: 100 ng for MATE1 and 200 ng for MATE2-K) and cellular uptake of [14C]TEA and [14C]metformin were reported earlier.11, 19, 21, 22
Uptake of [14C]TEA (tetraethylammonium) (a) and [14C]metformin (b) by HEK293 cells transiently expressing wild type or various multidrug and toxin extrusion 1 (MATE1) variants. The cells were preincubated with incubation medium (pH 7.4) in the presence of 30 mM ammonium chloride for 20 min. Then, the preincubation medium was removed, and the cells were incubated with 5 μM of [14C]TEA or 10 μM of [14C]metformin for 1 min at 37 °C. Each column represents the mean±s.d. of six monolayers from two independent experiments. **P<0.01, significantly different from the values for the wild type.
Uptake of [14C]TEA (tetraethylammonium) (a) and [14C]metformin (b) by HEK293 cells transiently expressing wild type or variants of multidrug and toxin extrusion 2-K (MATE2-K). The procedures are identical to those described in the legend of Figure 2. Each column represents the mean±s.d. of six monolayers from two independent experiments. **P<0.01, significantly different from the values for the wild type.
Concentration dependence of [14C]TEA (tetraethylammonium) uptake by HEK293 cells transiently expressing wild type or variants of multidrug and toxin extrusion 1 (MATE1) (a) and MATE2-K (b). The figures show a specific uptake of [14C]TEA obtained by subtracting the non-saturable components, which were estimated in the presence of 5 mM unlabeled compound. Each point represents the mean±s.d. of three monolayers from a typical experiment.
Cell surface biotinylation
Cell surface biotinylation was performed according to our earlier methods22 with some modifications. HEK293 cells were grown on poly-D-lysine-coated 12-well plates and transfected with MATE1 or MATE2-K cDNA plasmids (50 ng for MATE1 and 200 ng for MATE2-K). At 48 h after the transfection, cells were washed three times with 1 ml ice-cold phosphate-buffered saline with Ca and Mg (138 mM NaCl, 2.7 mM KCl, 1.5 mM KH2PO4, 9.6 mM Na2HPO4, 1 mM MgCl2 and 0.1 mM CaCl2, pH 7.3) and then treated with 400 μl of membrane-impermeable biotinylating agent, sulfo-NHS-SS-biotin (Pierce, Rockford, IL, USA) (1.5 mg ml−1) at 4 °C for 1 h. Subsequently, the cells were washed three times with 1 ml ice-cold phosphate-buffered saline with Ca and Mg containing 100 mM glycine and then incubated for 20 min at 4 °C with the same buffer to remove the remaining labeling agent. After being washed with phosphate-buffered saline with Ca and Mg, cells were disrupted with 400 μl of lysis buffer (10 mM Tris-base, 150 mM NaCl, 1 mM EDTA, 0.1% SDS, 1% Triton X-100 and 1% protease inhibitor cocktail (Nacalai Tesque, Kyoto, Japan), pH 7.4) at 4 °C for 1 h with constant agitation. Following centrifugation, 50 μl of streptavidin agarose beads (Pierce) was added to 300 μl of cell lysate and incubated for 1 h at room temperature to isolate the biotinylated membrane proteins.
Western blot analysis and quantification of band density
Isolated biotinylated membrane proteins were subjected to western blot analysis according to NuPAGE manufacturer's instructions (Invitrogen). Monoclonal anti-V5 antibody (Invitrogen) (1:2500 dilution) or Na+/K+-ATPase antibody (1:10 000 dilution; Upstate Biotechnology, Lake Placid, NY, USA) was used as the primary antibody. A peroxidase-conjugated anti-mouse IgG antibody was used for the detection of bound antibodies, and the blots were visualized by chemiluminescence on X-ray film. Quantification of band density was performed on scanned images using ImageJ, a public domain image-processing program (W Rasband, National Institute of Mental Health, Bethesda, MD, USA). The optical density of each lane was plotted, and the area under the curve was measured.
Statistical analysis
Kinetic parameter data were statistically analyzed with unpaired t-test compared with the values for the wild type. The other experimental data were statistically analyzed with the one-way analysis of variance followed by Dunnett's test.
Results
Identification of MATE1 and MATE2-K SNPs
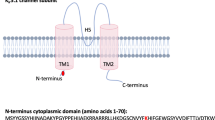
All 17 exons of the MATE1 and MATE2-K genes were sequenced to find SNPs in 89 Japanese subjects. In this study, eight MATE1 SNPs and five MATE2-K SNPs were identified in the coding region (Table 3). The allelic frequencies for the non-synonymous SNPs ranged from 0.6 to 2.2%. Figure 1 shows the position of mutated amino-acid residues in the predicted secondary structure of MATE1 (a) and MATE2-K (b), respectively. Only Ala310 was localized in the transmembrane domain (TMD), and other amino-acid residues are located at the intra- or extracellular domains.
Transport studies of the MATE1 and MATE2-K variants
To assess the functional alterations caused by the non-synonymous SNPs of both genes, [14C]TEA transport activity by the variants was evaluated by in vitro transient expression system. As shown in Figure 2a, [14C]TEA uptake by the MATE1 G64D variant was completely abolished. Other MATE1 variants except for the MATE1 V10L variant also showed a significant reduction in [14C]TEA transport activity, and the order of the remaining transport activities were as follows: wild type=V10L>N474S>D328A=A310V. [14C]Metformin uptake by various variants was similar to [14C]TEA uptake (Figure 2b). Both the MATE2-K variants showed significant decrease in [14C]TEA and [14C]metformin uptake, and the transport activity of MATE2-K G211V was completely abolished (Figures 3a and b).
Cell surface expression levels of the MATE1 and MATE2-K variants
To determine whether the reduced transport activity of these variants was due to the decreased expression of transporter proteins in the plasma membranes, cell surface biotinylation followed by western blot analysis was carried out. Among the MATE1 variants, the cell surface expression level of MATE1 G64D and D328A showed a decrease to approximately 10 and 20% compared with that of the wild-type MATE1 (Figure 4), which are well correlated with the reduction ratios of the transport activity for these variants (Figures 2a and b). Other MATE1 variants exhibited similar cell surface expression level with wild-type MATE1. In the MATE2-K, both the MATE2-K K64N and MATE2-K G211V variants showed a decrease to approximately 50 and 1% compared with that of the wild-type MATE2-K, respectively (Figure 5). These reduction ratios were well correlated with those of transport activities of both the MATE2-K variants (Figures 3a and b). These findings suggested that the low transport activities of MATE1 G64D, D328A and two MATE2-K variants were due to the alteration of protein expression in cell surface membranes.
Western blot analysis of cell surface biotinylated proteins obtained from HEK293 cells transiently expressing wild type or variants of multidrug and toxin extrusion 1 (MATE1). Cell surface membrane fractions prepared by cell surface biotinylation were separated by SDS-polyacrylamide gel electrophoresis (PAGE) (4–12%) and blotted onto polyvinylidene fluoride (PVDF) membranes. Each column represents the mean±s.d. of three monolayers. **P<0.01, significantly different from the values for the wild type.
Western blot analysis of cell surface biotinylated proteins obtained from HEK293 cells transiently expressing wild type or variants of multidrug and toxin extrusion 2-K (MATE2-K). The procedures are identical to those described in the legend of Figure 4. Each column represents the mean±s.d. of three monolayers. **P<0.01, significantly different from the values for the wild type.
Comparison of functional characteristics between wild type and the variants of MATE1 and MATE2-K
To estimate kinetic parameters for [14C]TEA uptake by several MATE1 and MATE2-K variants, concentration-dependent uptake was carried out (Figures 6a and b). The [14C]TEA uptake by the MATE1 and MATE2-K variants exhibited saturable kinetics, following the Michaelis–Menten equation. The apparent maximal uptake velocity (Vmax), Michaelis–Menten constant (Km) and Vmax/Km values are summarized in Table 4. Vmax values of MATE1 A310V, D328A and MATE2-K K64N were significantly decreased. Km values of MATE1 A310V and N474S were significantly increased.
Discussion
MATE1 and MATE2-K function as H+/organic cation antiporters at the renal brush border membranes and play crucial roles in the renal handling of cationic drugs, such as cimetidine, metformin and oxaliplatin.23 In this study, we identified five and two non-synonymous SNPs in their genes, all of which induced a reduction of transport activity except for MATE1 V10L. In MATE1, both the cell surface expression and transport activity of the G64D and D328A variants were significantly reduced to approximately 10 and 20% compared with that of the wild type. These findings suggest that reduced protein expression levels in the plasma membrane can account for the decrease in transport activity of MATE1 with G64D and D328A. Previously, we indicated that Cys63 of MATE1 plays an important role in substrate binding,22 and Cys63 is the neighboring amino-acid residue of Gly64. Thus, regarding G64D, the change from the small side chain (Gly) to the bulkier and polar side chain (Asp) may inhibit the substrate binding and decrease transport activities, in addition to reduced protein expression levels in the plasma membrane.
On the other hand, N474S sustained a modest level of transport activity. The small impact of Asn474 on MATE1 function may be due to its position, in the intracellular loop between TMD12 and TMD13. Three-dimensional models of MATE1 will clarify the importance of these amino-acid residues.
In the MATE1 protein, Gly64 and Asn474 are conserved in the rat (AB248823), mouse (AAH31436) and rabbit (EF120627) orthologs, suggesting that these two amino-acid residues are essential. Ala310 of MATE1 is conserved in the rat ortholog only. The transport activity of A310V showed a decrease to approximately 20% compared with that of the wild type, though its membrane expression level was same as the wild type. Kinetic analysis indicated that Km value for MATE1 A310V was significantly increased. These findings suggested that substitution of Ala for Val may inhibit the substrate binding or translocation because Ala310 was localized in the TMD.
In MATE2-K protein, Lys64 and Gly211 are conserved in the rabbit ortholog (EF121852). As for two MATE2-K SNPs, the alterations in transport activity were in accordance with the alterations in the cell surface expression of MATE2-K protein, indicating that the reduced function of MATE2-K K64N and MATE2-K G211V is mainly due to the decreased expression at the plasma membrane. Actually, it was demonstrated that Vmax value of MATE2-K K64N was significantly decreased. These findings may provide important information to elucidate molecular mechanisms of membrane trafficking and stability of the MATE2-K protein in the plasma membrane.
There are several reports that investigated the cell surface expression level caused by coding SNPs (cSNPs) between HEK293 cells and tissues. For example, the human organic anion-transporting polypeptide 1B1 protein expression level was not changed by substitution of Leu643 to Phe in both transfected HEK293 cells and liver samples.24 In this study, we examined the effect of cSNPs on cell surface expression level only in vitro. Our in vitro data are difficult to extrapolate to the case in vivo in renal proximal tubules; however, these data suggest that altered cell surface expression level may occur in individuals with MATE1 G64D, D328A and both the MATE2-K variants. Future studies on cell surface expression level of MATE1 and MATE2-K protein in human proximal tubules with these variants will elucidate whether in vitro data in this study are consistent with the case in vivo.
Metformin, a biguanide agent, is mainly excreted into the urine mostly through tubular secretion and shows large interindividual variation of renal clearance.18 Recently, it has been demonstrated that SNPs of the hepatic OCT1 gene in Caucasians and renal OCT2 gene in Koreans are responsible for the interindividual differences in the therapeutic efficacy and pharmacokinetics of metformin.15, 16, 17 On the other hand, Shikata et al.25 reported that OCT1 and OCT2 polymorphisms contribute little to the clinical efficacy of metformin in Japanese. Previously, we demonstrated that metformin is a good substrate not only for OCT2 but also for MATE1 and MATE2-K.19, 26 Therefore, the SNPs of MATE1 and MATE2-K genes identified in this study may be involved in the interindividual difference in the renal clearance of metformin in Japanese. However, as the allelic frequencies of MATE1 and MATE2-K SNPs are not very high, these SNPs cannot fully account for the large interindividual variation in the renal clearance of metformin.
We reported that the kidney-specific expression of OCT2 is involved in the renal distribution and accumulation of the anticancer agent cisplatin.27, 28 As there is little transport of cisplatin by MATE1 and MATE2-K, cisplatin is accumulated in the proximal tubular cells causing nephrotoxicity. A low-nephrotoxic platinum anticancer agent, oxaliplatin, was transported by OCT2 and MATE2-K,27, 28 suggesting that oxaliplatin does not accumulate in the renal proximal tubular cells. Therefore, loss of function of MATE2-K caused by cSNPs may lead to the accumulation of oxaliplatin in the kidney and the subsequent nephrotoxicity. Future study will be needed to clarify the clinical implications of the SNPs of both genes identified in this study.
In conclusion, five non-synonymous SNPs in the MATE1 and two non-synonymous SNPs in the MATE2-K genes were identified in Japanese subjects for the first time. All of the mutated proteins except for MATE1 V10L showed a significant decrease in transport activity, and especially those of MATE1 G64D and MATE2-K G211V were completely abolished by the impairment of cell surface expression. These polymorphisms may affect the renal handling of various cationic drugs and cause drug-induced nephrotoxicity.
Accession codes
References
Pritchard, J. B. & Miller, D. S. Mechanisms mediating renal secretion of organic anions and cations. Physiol. Rev. 73, 765–796 (1993).
Inui, K., Masuda, S. & Saito, H. Cellular and molecular aspects of drug transport in the kidney. Kidney Int. 58, 944–958 (2000).
Burckhardt, B. C. & Burckhardt, G. Transport of organic anions across the basolateral membrane of proximal tubule cells. Rev. Physiol. Biochem. Pharmacol. 146, 95–158 (2003).
Sekine, T., Miyazaki, H. & Endou, H. Molecular physiology of renal organic anion transporters. Am. J. Physiol. Renal Physiol. 290, F251–F261 (2006).
Koepsell, H., Lips, K. & Volk, C. Polyspecific organic cation transporters: structure, function, physiological roles, and biopharmaceutical implications. Pharm. Res. 24, 1227–1251 (2007).
Schuetz, J. D., Connelly, M. C., Sun, D., Paibir, S. G., Flynn, P. M., Srinivas, R. V. et al. MRP4: a previously unidentified factor in resistance to nucleoside-based antiviral drugs. Nat. Med. 5, 1048–1051 (1999).
Hasegawa, M., Kusuhara, H., Adachi, M., Schuetz, J. D., Takeuchi, K. & Sugiyama, Y. Multidrug resistance-associated protein 4 is involved in the urinary excretion of hydrochlorothiazide and furosemide. J. Am. Soc. Nephrol. 18, 37–45 (2007).
Ci, L., Kusuhara, H., Adachi, M., Schuetz, J. D., Takeuchi, K. & Sugiyama, Y. Involvement of MRP4 (ABCC4) in the luminal efflux of ceftizoxime and cefazolin in the kidney. Mol. Pharmacol. 71, 1591–1597 (2007).
Otsuka, M., Matsumoto, T., Morimoto, R., Arioka, S., Omote, H. & Moriyama, Y. A human transporter protein that mediates the final excretion step for toxic organic cations. Proc. Natl Acad. Sci. USA 102, 17923–17928 (2005).
Masuda, S., Terada, T., Yonezawa, A., Tanihara, Y., Kishimoto, K., Katsura, T. et al. Identification and functional characterization of a new human kidney-specific H+/organic cation antiporter, kidney-specific multidrug and toxin extrusion 2. J. Am. Soc. Nephrol. 17, 2127–2135 (2006).
Tsuda, M., Terada, T., Asaka, J., Ueba, M., Katsura, T. & Inui, K. Oppositely directed H+ gradient functions as a driving force of rat H+/organic cation antiporter MATE1. Am. J. Physiol. Renal Physiol. 29, F593–F598 (2007).
Ingelman-Sundberg, M., Sim, S. C., Gomez, A. & Rodriguez-Antona, C. Influence of cytochrome P450 polymorphisms on drug therapies: pharmacogenetic, pharmacoepigenetic and clinical aspects. Pharmacol. Ther. 116, 496–526 (2007).
Ishikawa, T., Tsuji, A., Inui, K., Sai, Y., Anzai, N., Wada, M. et al. The genetic polymorphism of drug transporters: functional analysis approaches. Pharmacogenomics 5, 67–99 (2004).
Kivistö, K. T. & Niemi, M. Influence of drug transporter polymorphisms on pravastatin pharmacokinetics in humans. Pharm. Res. 24, 239–247 (2007).
Shu, Y., Brown, C., Castro, R. A., Shi, R. J., Lin, E. T., Owen, R. P. et al. Effect of genetic variation in the organic cation transporter 1, OCT1, on metformin pharmacokinetics. Clin. Pharmacol. Ther. 83, 273–280 (2008).
Shu, Y., Sheardown, S. A., Brown, C., Owen, R. P., Zhang, S., Castro, R. A. et al. Effect of genetic variation in the organic cation transporter 1 (OCT1) on metformin action. J. Clin. Invest. 117, 1422–1431 (2007).
Song, I. S., Shin, H. J., Shim, E. J., Jung, I. S., Kim, W. Y., Shon, J. H. et al. Genetic variants of the organic cation transporter 2 influence the disposition of metformin. Clin. Pharmacol. Ther. 84, 559–562 (2008).
Yin, O. Q., Tomlinson, B. & Chow, M. S. Variability in renal clearance of substrates for renal transporters in Chinese subjects. J. Clin. Pharmacol. 46, 157–163 (2006).
Tanihara, Y., Masuda, S., Sato, T., Katsura, T., Ogawa, O. & Inui, K. Substrate specificity of MATE1 and MATE2-K, human multidrug and toxin extrusions/H+-organic cation antiporters. Biochem. Pharmacol. 74, 359–371 (2007).
Kajiwara, M., Terada, T., Asaka, J., Ogasawara, K., Katsura, T., Ogawa, O. et al. Critical roles of Sp1 in gene expression of human and rat H+/organic cation antiporter MATE1. Am. J. Physiol. Renal Physiol. 293, F1564–F1570 (2007).
Terada, T., Masuda, S., Asaka, J., Tsuda, M., Katsura, T. & Inui, K. Molecular cloning, functional characterization and tissue distribution of rat H+/organic cation antiporter MATE1. Pharm. Res. 23, 1696–1701 (2006).
Asaka, J., Terada, T., Tsuda, M., Katsura, T. & Inui, K. Identification of essential histidine and cysteine residues of the H+/organic cation antiporter multidrug and toxin extrusion (MATE). Mol. Pharmacol. 71, 1487–1493 (2007).
Terada, T. & Inui, K. Physiological and pharmacokinetic roles of H+/organic cation antiporters (MATE/SLC47A). Biochem. Pharmacol. 75, 1689–1696 (2008).
Seithel, A., Klein, K., Zanger, U. M., Fromm, M. F. & König, J. Non-synonymous polymorphisms in the human SLCO1B1 gene: an in vitro analysis of SNP c.1929A>C. Mol. Genet. Genomics 279, 149–157 (2008).
Shikata, E., Yamamoto, R., Takane, H., Shigemasa, C., Ikeda, T., Otsubo, K. et al. Human organic cation transporter (OCT1 and OCT2) gene polymorphisms and therapeutic effects of metformin. J. Hum. Genet. 52, 117–122 (2007).
Kimura, N., Masuda, S., Tanihara, Y., Ueo, H., Okuda, M., Katsura, T. et al. Metformin is a superior substrate for renal organic cation transporter OCT2 rather than hepatic OCT1. Drug Metab. Pharmacokinet. 20, 379–386 (2005).
Yokoo, S., Yonezawa, A., Masuda, S., Fukatsu, A., Katsura, T. & Inui, K. Differential contribution of organic cation transporters, OCT2 and MATE1, in platinum agent-induced nephrotoxicity. Biochem. Pharmacol. 74, 477–487 (2007).
Yonezawa, A., Masuda, S., Yokoo, S., Katsura, T. & Inui, K. Cisplatin and oxaliplatin, but not carboplatin and nedaplatin, are substrates for human organic cation transporters (SLC22A1-3 and multidrug and toxin extrusion family). J. Pharmacol. Exp. Ther. 319, 879–886 (2006).
Acknowledgements
This study was supported, in part, by a Grant-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology of Japan, and a Grant-in-Aid for Research on Advanced Medical Technology from the Ministry of Health, Labor and Welfare of Japan. KO is a Research Fellow of the Japan Society for the Promotion of Science.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Kajiwara, M., Terada, T., Ogasawara, K. et al. Identification of multidrug and toxin extrusion (MATE1 and MATE2-K) variants with complete loss of transport activity. J Hum Genet 54, 40–46 (2009). https://doi.org/10.1038/jhg.2008.1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/jhg.2008.1
Keywords
This article is cited by
-
Prevention of Cisplatin-Induced Acute Kidney Injury: A Systematic Review and Meta-Analysis
Drugs (2019)
-
Renal Drug Transporters and Drug Interactions
Clinical Pharmacokinetics (2017)
-
Structure and function of multidrug and toxin extrusion proteins (MATEs) and their relevance to drug therapy and personalized medicine
Archives of Toxicology (2016)
-
Association of decreased mRNA expression of multidrug and toxin extrusion protein 1 in peripheral blood cells with the development of flutamide-induced liver injury
Cancer Chemotherapy and Pharmacology (2015)
-
Individualized Therapy for Type 2 Diabetes
Molecular Diagnosis & Therapy (2012)