Abstract
Bone mineral density (BMD), the major factor determining bone strength, is closely related to osteoporotic fracture risk and is determined largely by multiple genetic factors. Semaphorin 7a (SEMA7A), a recently described member of the semaphorin family, has been shown to play a critical role in the activation of monocyte/macrophages that share progenitors with bone-resorbing osteoclasts and thus might contribute to osteoclast development. In the present study, we directly sequenced the SEMA7A gene in 24 Korean individuals, and identified 15 sequence variants. Five polymorphisms (+15667G>A, +15775C>G, +16285C>T, +19317C>T, +22331A>G) were selected and genotyped in postmenopausal Korean women (n=560) together with measurement of the areal BMD (g/cm2) of the anterior-posterior lumbar spine and the non-dominant proximal femur using dual-energy X-ray absorptiometry. We found that polymorphisms of the SEMA7A gene were associated with the BMD of the lumbar spine and femoral neck. SEMA7A+15775C>G and SEMA7A+22331A>G were associated with low BMD of the femoral neck (P=0.02) and lumbar spine (P=0.04) in a recessive model. SEMA7A-ht4 also showed an association with risk of vertebral fracture (OR=1.87–1.93, P=0.02–0.03). Our results suggest that variations in SEMA7A may play a role in decreased BMD and risk of vertebral fracture.
Similar content being viewed by others
Introduction
Osteoporosis is a systemic bone disease characterized by low bone mineral density (BMD) and increased risk of fracture (Peacock et al. 2002). Although multiple risk factors influence BMD and thus the pathogenesis of osteoporosis, genetic factors are mainly implicated, accounting for about 50–85% of the variance in BMD based on twin and family studies (Slemenda et al. 1991; Kanis et al. 1994; Lander and Schork 1994; Weeks and Lathrop 1995; Arden and Spector 1997; Eisman 1999; Stewart and Ralston 2000). In a pathological condition, an imbalance of bone remodeling occurs due to the increased rate of bone resorption by osteoclasts over the rate of bone formation by osteoblasts. Therefore, genes involved in osteoclast differentiation and function are potential candidate genes for having a genetic effect on BMD and osteoporosis.
Semaphorin7A (SEMA7A), located on chromosome 15q22.3-q23, has been implicated in the regulation of the immune and nervous systems (Comeau et al. 1998; Lange et al. 1998; Xu et al. 1998; Pasterkamp et al. 2003). SEMA7A is a cell surface protein associated through a glycophosphatidylinositol (GPI) linkage (Xu et al. 1998), and has been shown to be a potent activator of macrophages. SEMA7A induces chemotaxis, inflammatory cytokine production [interleukin-1 (IL-1β), tumour necrosis factor-α (TNF-α), IL-6 and IL-8] and superoxide release in monocytes whereas it does not influence cytokine production in B and T cells (Holmes et al. 2002). SEMA7A was also shown to induce the production of granulocyte-macrophage colony stimulating factor (GM-CSF) and the differentiation of monocytes into dendritic cells (Holmes et al. 2002). Recently it was shown that SEMA7A is capable of increasing the migration of MC3T3 cells and enhancing the fusion of osteoclasts (Delorme et al. 2005). These results suggest that SEMA7A plays a critical role in the differentiation and activation of myeloid cells such as macrophages and osteoclasts.
In the present study, we investigated the genetic effects of SEMA7A on the determination of bone mass by direct resequencing of the gene to detect polymorphisms, and by analyzing its association with BMD and fracture risk in postmenopausal Korean women.
Materials and methods
Subjects
The study population, comprising 560 postmenopausal women of Korean ethnicity who visited Asan Medical Center (AMC, Seoul, Korea), was previously reported (Kim et al. 2005). Menopause was defined as the absence of menstruation for at least 6 months, and was confirmed by measurement of serum follicle-stimulating hormone (FSH). Women with premature menopause (before 40 years of age) were excluded. Women who had taken drugs that might affect bone metabolism for more than 6 months or within the previous 12 months were also excluded. Subjects were excluded if they had suffered from any disease that might affect bone metabolism. Women who had had a stroke or dementia were also excluded because of concerns related to their limited physical activity. Women were also excluded if they had osteophyte formation above the fourth grade of the Nathan classification (Nathan 1962), and/or severe facet joint osteoarthritis in the lumbar spine diagnosed using conventional spine radiographs. The study was approved by the AMC ethics review committee and written informed consent was obtained from all subjects.
Areal BMD (g/cm2) of the anterior-posterior lumbar spine (L2–L4) and proximal femur (femoral neck, femoral shaft, total femur, trochanter and Ward’s triangle) was measured using dual energy X-ray absorptiometry (Lunar Expert XL with software version 1.90; Madison, WI) in 431 women. In 129 women, BMD was measured using Hologic QDR 4500-A with software version 4.84 (Waltham, MA). The Hologic machine did not measure BMD at the femoral shaft, thus BMD values were measured only at the lumbar spine and femoral neck before January 2001 in our institution, i.e., BMD values at the femoral shaft and at other proximal femur sites (total femur, trochanter and Ward’s triangle) were not available in 229 and 100 subjects, respectively. Because of upper extremity dominance, the BMD at the proximal femur was measured at non-dominant sites. Short-term in vivo measurement precision for the Lunar and Hologic machines, expressed as the coefficient of variation, were 0.82 and 0.85% for the lumbar spine, respectively, and 1.12 and 1.20% for the femoral neck, respectively. These values were obtained by scanning 17 volunteers who were not part of the study; each volunteer underwent five scans on the same day, getting on and off the table between examinations. To derive cross-calibration equations between the two systems, BMD values were measured at the lumbar and femoral neck by the two machines in 109 healthy Korean women (55±11 years, range 31–75 years), and cross-calibration equations were calculated as follows (Jo et al. 1999):
-
L2–L4 BMD (g/cm2): Lunar = 1.1287 × Hologic − 0.0027
-
Femoral neck BMD (g/cm2): Lunar = 1.1556 × Hologic + 0.0182
The T-score was used to diagnose osteopenia and osteoporosis at each site according to the World Health Organization (WHO) definition (−2.5 < T-score < −1.0 SD and T-score ≤ −2.5 SD, respectively); subjects having a T-score over −1.0 were classified as normal controls. Lateral thoracolumbar (T4–L4) radiographs were obtained for all subjects. A vertebral fracture was defined quantitatively as a loss of 15% or more in the anterior, posterior, or middle height of one or more vertebral sites in subjects without previous history of major trauma such as traffic accident.
Sequencing analysis of the SEMA7A gene
Genomic DNA was extracted from peripheral blood leukocytes using a commercial kit (Wizard Genomic DNA purification kit; Promega, Madison, WI). We sequenced exons and their boundaries of the SEMA7A gene, including the promoter region (~1.5 kb), to discover genetic variants in 24 Korean DNA samples using a DNA analyzer (ABI PRISM 3700; Applied Biosystems, Foster City, CA). Fourteen primer sets for the SEMA7A gene for amplification and sequencing analysis were designed based on GenBank sequences (reference genomic sequence for SEMA7A: accession number NM_003612). Information regarding the primers used is available on our website; sequence variants were verified by chromatograms (http://www.snp-genetics.com/reference/SEMA7A_add_info.doc).
Genotyping using fluorescence polarization detection
For genotyping of polymorphic sites, amplifying primers and probes were designed for TaqMan (Livak 1999). Primer Express (Applied Biosystems) was used to design both the PCR primers and the MGB TaqMan probes. One allelic probe was labeled with FAM dye and the other with the fluorescent dye VIC. PCR was performed using TaqMan Universal Master mix without UNG (Applied Biosystems), with PCR primer concentrations of 900 nM and TaqMan MGB-probe concentrations of 200 nM. Reactions were performed in a 384-well format in a total reaction volume of 5 μl using 20 ng genomic DNA. The plates then were placed in a thermal cycler (PE 9700, Applied Biosystems) and heated at 50°C for 2 min and 95°C for 10 min, followed by 40 cycles of 95°C for 15 s and 60°C for 1 min. The TaqMan assay plates were transferred to a Prism 7900HT instrument (Applied Biosystems) where the fluorescence intensity in each well of the plate was read. Fluorescence data files from each plate were analyzed using automated software (SDS 2.1, Applied Biosystems). Information regarding the primers is available on our website (http://www.snp-genetics.com/reference/SEMA7A_add_info.doc).
Statistics
To determine whether individual variants were in equilibrium at each locus in the population (Hardy–Weinberg equilibrium), χ2tests were applied. We examined Lewontin’s D′ (|D′|) and the linkage disequilibrium (LD) coefficient, r2, between all pairs of biallelic loci (Hedrick 1987; Hedrick and Kumar 2001). Haplotypes (ht) of each individual were inferred using the algorithm developed by Stephens et al. (2001) (PHASE), which uses a Bayesian approach incorporating a priori expectations of haplotypic structure based on population genetics and coalescent theory. Phase probabilities of all polymorphic sites for haplotypes were calculated for each individual by this software. Individuals with phase probabilities less than 97% were excluded from the analysis. The genetic effects of inferred haplotypes were analyzed in the same way as polymorphisms. Multiple regression analyses were performed for BMD, controlling for age (continuous variable), years since menopause (YSM; continuous variable), weight, height, and type of evaluation machine as covariants. The genotype distributions of SEMA7A polymorphisms and haplotypes between patient groups, including osteopenic and osteoporotic subjects and normal subjects, were analyzed with logistic regression models controlling for age, YSM, weight, height, and type of evaluation machine as covariants. In addition, genotype and haplotype distributions between patients with and without vertebral fractures were also analyzed with logistic regression model controlling for age, YSM, weight and height.
Results and discussion
The mean age of the participants was 59.4±7.2 years (range 46–83 years), and the mean YSM was 10.4±8.2 years (range 1–35 years) (Table 1). As expected, age, weight, height, and YSM were significantly correlated with BMD at the lumbar spine and femoral neck, with the exception of the correlation between femoral neck BMD and height (Table 1). BMDs measured by Lunar equipment (0.870±0.182 and 0.723±0.128 g/cm2 at the lumbar spine and femoral neck, respectively) were significantly higher than those measured by Hologic equipment (0.764±0.116 and 0.606 ± 0.098 g/cm2, respectively; P < 0.0001 for both). Therefore, cross-calibration of BMD in the spine and femoral neck was carried out as described in Materials and methods.
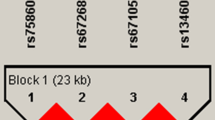
Although SEMA7A has been implicated in the activation of monocytes/macrophages and in the induction of osteoclast fusion, no information about its genetic effects on human disease is available. To identify polymorphisms in SEMA7A, we directly sequenced all exons and their boundaries of the SEMA7A gene, including −1,500 bp of the 5′ flanking region, and identified fifteen polymorphisms (one in the 5′ flanking region, eight in exons and six in introns) in a Korean population. Of the 15 polymorphisms identified, 6 (−1396A>G, +15667G>A, +15955C>T C126C, +16412C>G, +16855C>G, and +18986G>A V334I) were newly identified in this study; their frequencies are shown in Fig. 1a. Genotype distributions of all loci were in Hardy–Weinberg equilibrium (P>0.05). We selected five polymorphisms (+15667G>A, +15775C>G, +16285C>T, +19317C>T, and +22331A>G) for a larger scale genotyping (n=560) by considering their position, allele frequencies, haplotype-tagging status, and LD coefficients among polymorphisms. SEMA7A gene haplotypes were constructed using PHASE software (Stephens et al. 2001) (Fig. 1b).
Gene maps and haplotypes of SEMA7A. Black blocks Coding exons; white blocks 5′ and 3′ UTRs, +1 first base of translation start site; asterisks polymorphisms genotyped in a larger population (n=557). The frequencies of polymorphisms not subjected to larger scale genotyping were based on sequence data (n=24). a Polymorphisms identified in SEMA7A on chromosome 15q22.3-q23 (reference genome sequence: NT_010194). b Haplotypes of SEMA7A. Only those with frequencies over 0.01 are shown; others* includes rare haplotypes: GCTCG, GCCTG, GGCCG, ACTCA, and GGTCG. c Linkage disequilibrium (LD) coefficients (|D′| and r2) among SEMA7A polymorphisms
Using multiple regression analysis for association of BMD with lumbar spine, femoral neck and other bone sites, controlling for age, YSM, height, weight, and evaluation machines as covariants, SEMA7A+22331A>G and SEMA7A+15775C>G were associated with low BMD of lumbar spine (P=0.04) and femoral neck (P=0.02), respectively (Table 2). The BMD levels of lumbar spine (L2–L4) in individuals bearing minor homozygous genotype [0.85±0.16 (G/G of +22331A>G)] were lower than those in individuals bearing common homozygous and heterozygous genotypes (0.88±0.17 and 0.88±0.17, respectively; P=0.04, Table 2). Similarly, BMD levels of the femoral neck in individuals bearing minor homozygous genotype [0.68±0.13 (G/G of +15775C>G)] were lower than those in individuals bearing common homozygous and heterozygous genotypes (0.72±0.12 and 0.73±0.12, respectively; P=0.02, Table 2). Construction of haplotypes (Fig. 1b) and subsequent analysis revealed that SEMA7A-ht4 (ACTTG) was associated with susceptibility to risk of fracture (OR=1.87–1.93, P=0.02–0.03; Table 3). The frequency of SEMA7A-ht4 was higher in the osteoporosis group than in that of the normal control. This haplotype included the +22331G allele, which was associated with low BMD at the lumbar spine, suggesting that SEMA7A+22331A>G contributes to both low BMD at the lumbar spine and high risk of vertebral fracture.
Recently, SEMA7A was shown to increase the migration of MC3T3 osteoblasts and to enhance the fusion of osteoclasts (Delorme et al. 2005), suggesting a critical role in bone cells. Consistently, our association study of SEMA7A polymorphism with BMD and risk of fracture demonstrates an association of SEMA7A polymorphisms with low BMD of lumbar spine and femoral neck. SEMA7A-ht4 was also associated with high risk of vertebral fracture. To our knowledge, this is the first clinical report suggesting a role for SEMA7A in bone metabolism. Although the genetic effects of SEMA7A polymorphism on BMD and fracture risk are not dramatic, i.e., associated P-values did not retain significance after correction of multiple comparisons, it might be worthwhile to follow up on the signals of this important gene through larger cohort studies. Further biological and/or functional evidence would be needed to confirm the suggestive association of SEMA7A polymorphism with BMD.
In summary, to examine the possible involvement of genetic polymorphisms of SEMA7A in osteoporosis, 15 polymorphisms in SEMA7A were identified [nine known and six novel single nucleotide polymorphisms (SNPs)] and five SNPs were genotyped in postmenopausal Korean women (n=560). Statistical analyses found genetic linkage of SEMA7A+15775C>G and SEMA7A+22331A>G with decreased BMD of spine and femoral neck as well as linkage of SEMA7A-ht4 with risk of vertebral fracture in Korean postmenopausal women.
References
Arden NK, Spector TD (1997) Genetic influences on muscle strength, lean body mass, and bone mineral density: a twin study. J Bone Miner Res 12:2076–2081
Comeau MR, Johnson R, DuBose RF, Petersen M, Gearing P, VandenBos T, Park L, Farrah T, Buller RM, Cohen JI, Strockbine LD, Rauch C, Spriggs MK (1998) A poxvirus-encoded semaphorin induces cytokine production from monocytes and binds to a novel cellular semaphorin receptor, VESPR. Immunity 8:473–482
Delorme G, Saltel F, Bonnelye E, Jurdic P, Machuca-Gayet I (2005) Expression and function of semaphorin 7A in bone cells. Biol Cell 97:589–597
Eisman JA (1999) Genetics of osteoporosis. Endocr Rev 20:788–804
Hedrick P, Kumar S (2001) Mutation and linkage disequilibrium in human mtDNA. Eur J Hum Genet 9:969–972
Hedrick PW (1987) Gametic disequilibrium measures: proceed with caution. Genetics 117:331–341
Holmes S, Downs AM, Fosberry A, Hayes PD, Michalovich D, Murdoch P, Moores K, Fox J, Deen K, Pettman G, Wattam T, Lewis C (2002) Sema7A is a potent monocyte stimulator. Scand J Immunol 56:270–275
Jo JM, Kim JS, Kim GS, Kim SW, Shin JW, Moon DH, Lee HK (1999) Cross-calibration of bone mineral density between two different dual X-ray absorptiometry systems: hologic QDR 4500-A and lunar EXPERT-XL. Korean J Nucl Med 33:282–288
Kanis JA, Melton LJ III, Christiansen C, Johnston CC, Khaltaev N (1994) The diagnosis of osteoporosis. J Bone Miner Res 9:1137–1141
Kim GS, Koh JM, Chang JS, Park BL, Kim LH, Park EK, Kim SY, Shin HD (2005) Association of the OSCAR promoter polymorphism with BMD in postmenopausal women. J Bone Miner Res 20:1342–1348
Lander ES, Schork NJ (1994) Genetic dissection of complex traits. Science 265:2037–2048
Lange C, Liehr T, Goen M, Gebhart E, Fleckenstein B, Ensser A (1998) New eukaryotic semaphorins with close homology to semaphorins of DNA viruses. Genomics 51:340–350
Livak KJ (1999) Allelic discrimination using fluorogenic probes and the 5′ nuclease assay. Genet Anal 14:143–149
Nathan H (1962) Osteophytes of the vertebral column: an anatomical study of their development according to age, race, and sex with considerations as to their etiology and significance. J Bone Joint Surg Am 44A:243–268
Pasterkamp RJ, Peschon JJ, Spriggs MK, Kolodkin AL (2003) Semaphorin 7A promotes axon outgrowth through integrins and MAPKs. Nature 424:398–405
Peacock M, Turner CH, Econs MJ, Foroud T (2002) Genetics of osteoporosis. Endocr Rev 23:303–326
Slemenda CW, Christian JC, Williams CJ, Norton JA, Johnston CC Jr (1991) Genetic determinants of bone mass in adult women: a reevaluation of the twin model and the potential importance of gene interaction on heritability estimates. J Bone Miner Res 6:561–567
Stephens M, Smith NJ, Donnelly P (2001) A new statistical method for haplotype reconstruction from population data. Am J Hum Genet 68:978–989
Stewart TL, Ralston SH (2000) Role of genetic factors in the pathogenesis of osteoporosis. J Endocrinol 166:235–245
Weeks DE, Lathrop GM (1995) Polygenic disease: methods for mapping complex disease traits. Trends Genet 11:513–519
Xu X, Ng S, Wu ZL, Nguyen D, Homburger S, Seidel-Dugan C, Ebens A, Luo Y (1998) Human semaphorin K1 is glycosylphosphatidylinositol-linked and defines a new subfamily of viral-related semaphorins. J Biol Chem 273:22428–22434
Acknowledgement
This study was supported by a grant from the Korea Health 21 R&D Project, Ministry of Health & Welfare, Republic of Korea (Project No.: 01-PJ3-PG6-01GN11-0002).
Author information
Authors and Affiliations
Corresponding author
Additional information
Jung-Min Koh and Bermseok Oh contributed equally to this work
Electronic Supplementary Material
Rights and permissions
About this article
Cite this article
Koh, JM., Oh, B., Lee, J.Y. et al. Association study of semaphorin 7a (sema7a) polymorphisms with bone mineral density and fracture risk in postmenopausal Korean women. J Hum Genet 51, 112–117 (2006). https://doi.org/10.1007/s10038-005-0331-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10038-005-0331-z
Keywords
This article is cited by
-
Semaphorin 7a exerts pleiotropic effects to promote breast tumor progression
Oncogene (2016)
-
Bone cell communication factors and Semaphorins
BoneKEy Reports (2012)
-
Runx2 promotes both osteoblastogenesis and novel osteoclastogenic signals in ST2 mesenchymal progenitor cells
Osteoporosis International (2012)
-
Suppression of bone formation by osteoclastic expression of semaphorin 4D
Nature Medicine (2011)
-
Plexin C1, A Receptor for Semaphorin 7A, Inactivates Cofilin and Is a Potential Tumor Suppressor for Melanoma Progression
Journal of Investigative Dermatology (2009)