Abstract
We identified a total of 187 single-nucleotide polymorphisms (SNPs) at 11 gene loci in the 130-kb region on chromosome 6p21 containing a gene strongly associated with myocardial infarction (MI). By comparing our data with SNPs deposited in the dbSNP database at the National Center for Biotechnology Information, 46 of these SNPs (24.6%) were considered to be novel: four were identified in the P5-1 locus, 14 in the MICB locus, nine in the BAT1 locus, one in the ATP6V1G2 locus, six in the NFKBIL1 locus, one in the LTA locus, one in the TNF locus, five in the LST1 locus, four in the LY117a locus, and one in the AIF-1 locus. The SNP map presented here should provide as useful resource not only for examining the relationships between genotypes and susceptibility to the MI phenotype, but also for scanning of complex diseases mapped to this local segment on chromosome 6.
Similar content being viewed by others
Introduction
Despite the changes in life style and development of new pharmacologic approaches, coronary artery diseases including myocardial infarction (MI) remain the principal causes of death in the world, indicating the importance of identifying genetic and environmental factors in their pathogenesis (for reviews, see Braunwald 1997; Breslow 1997). By means of a genome-wide gene-based single-nucleotide polymorphism (SNP) analysis and subsequent functional assay, we previously identified the LTA gene as an MI susceptibility gene on 6p21, and two genetic variants in the gene that may influence lymphotoxin-alpha (LTA) function both quantitatively and qualitatively (Ozaki et al. 2002). During the search for the MI susceptibility gene in this critical region, we attempted to construct a high-density SNP map for linkage disequilibrium (LD) mapping by direct sequencing analysis and identified one extended block of intense LD with LD coefficients drop-off near P5-1 and AIF-1 (Ozaki et al. 2002).
In this paper, we provide a high-resolution SNP map of this MI critical region on 6p21, in which we have detected a total of 46 novel SNPs among 64 chromosomes from 16 individuals with MI and 16 control individuals.
Subjects and methods
Blood samples were obtained with written informed consent from 16 individuals with MI and 16 control individuals for this study, which was approved by the ethical committee of the RIKEN SNP Research Center. We generated a reference sequence of 130-kb by assembling sequences Y14768, AP000506, and AC004184 from the GenBank database and then designed primer sets to amplify the 11 gene loci (AIF-1, LY117a, LST1, LTB, TNF, LTA, NFKBIL1, ATP6V1G2, BAT1, MICB, and P5-1), excluding over most of the regions that corresponded to repetitive sequences predicted by the Repeat Masker program (http://repeatmasker.genome.washington.edu/cgi-bin/RepeatMasker).
Each polymerase chain reaction (PCR) was performed with 20 ng mixed genomic DNA derived from two individuals. Amplification of genomic DNA fragments by PCR, and DNA sequencing of the amplified fragments were performed according to methods described previously (Iida et al. 2003).
Results and discussion
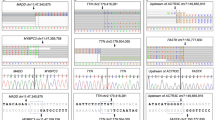
By direct sequencing of DNA from 16 individuals with MI and 16 control individuals, we screened SNPs in a 130-kb genomic region corresponding to the MI critical region on 6p21, which contains 11 genes, except for most of the regions of human repetitive sequences, and identified 187 SNPs. SNPs were distributed every 275 nucleotides on average in the 51.4-kb regions screened, although their densities varied considerably in these regions. We found 68 SNPs in the MICB gene locus of which 8.6-kb was examined (1 SNP/127 nucleotides), whereas only two were identified in the 3.6-kb genomic region containing the LTB gene (1 SNP/1783 nucleotides). By a comparison of our data with SNPs deposited in the dbSNP database in National Center for Biotechnology Information (NCBI 2003), 46 of the 187 SNPs were considered to be novel (Table 1). Of the 46 SNPs, four were identified in the P5-1 gene locus, 14 in the MICB gene locus, nine in the BAT1 gene locus, one in the ATP6V1G2 gene locus, six in the NFKBIL1 gene locus, one in the LTA gene locus, one in the TNF gene locus, five in the LST1 gene locus, four in the LY117a gene locus, and one in the AIF-1 gene locus (Fig. 1, Table 1). The SNPs consisted of 32 transitions and 14 transversions; thus, transitions occurred 2.3 times more frequently than transversions.
Transcription map and locations of 46 novel single nucleotide polymorphisms (SNPs) in the 130-kb region containing a myocardial infarction susceptibility gene on chromosomal band 6p21. Each gene and its orientation are represented by an open rectangle and an arrowhead, respectively. SNPs are indicated below each gene as vertical lines
The gene-based SNP map presented here should provide as useful resource not only for further examining the relationships between genotypes and susceptibility to the MI phenotype, but also for screening of genes associated with complex diseases mapped to this local segment on chromosome 6.
References
Braunwald E (1997) Shattuck lecture—cardiovascular medicine at the turn of the millennium: triumphs, concerns, and opportunities. N Engl J Med 337:1360–1369
Braunwald E (1997) Shattuck lecture—cardiovascular medicine at the turn of the millennium: triumphs, concerns, and opportunities. N Engl J Med 337:1360–1369Breslow JL (1997) Cardiovascular disease burden increases, NIH funding decreases. Nature Med 3:600–601
Braunwald E (1997) Shattuck lecture—cardiovascular medicine at the turn of the millennium: triumphs, concerns, and opportunities. N Engl J Med 337:1360–1369Breslow JL (1997) Cardiovascular disease burden increases, NIH funding decreases. Nature Med 3:600–601Dunnen JT den, Antonarakis SE (2000) Mutation nomenclature extensions and suggestions to describe complex mutations: a discussion. Hum Mutat 15:7–12
Braunwald E (1997) Shattuck lecture—cardiovascular medicine at the turn of the millennium: triumphs, concerns, and opportunities. N Engl J Med 337:1360–1369Breslow JL (1997) Cardiovascular disease burden increases, NIH funding decreases. Nature Med 3:600–601Dunnen JT den, Antonarakis SE (2000) Mutation nomenclature extensions and suggestions to describe complex mutations: a discussion. Hum Mutat 15:7–12Iida A, Saito S, Sekine A, Mishima C, Kitamura Y, Kondo K, Harigae S, Osawa S, Nakamura Y (2003) Catalog of 668 SNPs detected among 31 genes encoding potential drug targets on the cell surface. J Hum Genet 48:23–46
Braunwald E (1997) Shattuck lecture—cardiovascular medicine at the turn of the millennium: triumphs, concerns, and opportunities. N Engl J Med 337:1360–1369Breslow JL (1997) Cardiovascular disease burden increases, NIH funding decreases. Nature Med 3:600–601Dunnen JT den, Antonarakis SE (2000) Mutation nomenclature extensions and suggestions to describe complex mutations: a discussion. Hum Mutat 15:7–12Iida A, Saito S, Sekine A, Mishima C, Kitamura Y, Kondo K, Harigae S, Osawa S, Nakamura Y (2003) Catalog of 668 SNPs detected among 31 genes encoding potential drug targets on the cell surface. J Hum Genet 48:23–46NCBI (2003) dbSNP database (June 2003), http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=snp
Ozaki K, Ohnishi Y, Iida A, Sekine A, Yamada R, Tsunoda T, Sato H, Sato H, Hori M, Nakamura Y, Tanaka T (2002) Functional SNPs in the lymphotoxin-alpha gene that are associated with susceptibility to myocardial infarction. Nat Genet 32:650–654
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Iida, A., Ozaki, K., Ohnishi, Y. et al. Identification of 46 novel SNPs in the 130-kb region containing a myocardial infarction susceptibility gene on chromosomal band 6p21. J Hum Genet 48, 476–479 (2003). https://doi.org/10.1007/s10038-003-0054-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10038-003-0054-y