Abstract
Ichthyosis vulgaris (IV) is an inherited scaling skin disorder with a prevalence estimated at 2.29% in China. The gene responsible for this disorder has not been elucidated. To find the disease gene, we ascertained two Chinese IV families. Linkage analysis identified an IV locus on chromosome 1q22 with a maximum two-point Lod score of 2.47 at D1S1653 (θ=0.00). Haplotype analysis placed the critical region in a 7-cM interval defined by D1S1653 and D1S2675. These results provide the basis for further identifying the gene responsible for IV disorder.
Similar content being viewed by others
Introduction
Ichthyosis vulgaris (IV) is the common form of inherited scaling disorder, with a prevalence estimated at 1 in 250–5,300 (Wells and Kerr 1966). In the Chinese population, the prevalence is about 2.29% (Lei et al. 1992). The onset of IV is usually within one year after birth. The patients manifest with white scaling on the extensor surfaces of the extremities and occasionally the trunk. Clinical features also include hyperlinearity of the palms and soles, atopy and heat intolerance or keratosis pilaris. Genetically, IV is thought to be an autosomal dominant disorder with virtually complete penetrance (Wells and Kerr 1966).
Although the symptoms of X-linked ichthyosis (XLI) are similar to IV, the format of the scales and involvement of the preauricular area were found to be reliable signs to distinguish them. In XLI patients, scales are mostly large and dark brown, in contrast, small and light-brown scales are seen in IV patients. The presence of lesions was found in 93% of the cases with XLI, while only 17% of the IV patients had ichthyotic lesions at this site (Okano et al.1988). Genetically, the steroid sulphatase gene (STS) responsible for XLI has been mapped to the distal part of the short arm of the X chromosome (Hernandez-Martin et al. 1999).
Due to the difficulties in collecting suitable large affected families, linkage studies to map the gene involved in IV are rarely done. Recently, two multigenerational families with affected individuals showing clinical features of IV but distinct histological features were studied by gene linkage, and the results suggested the epidermal differentiation complex on chromosome 1 might be involved (Compton et al. 2002). Furthermore, strong evidence revealed the crucial importance of filaggrin (encoded by FLG) in normal epidermal function and the molecular basis of IV (Nirunsuksiri et al.1995; Presland et al. 2000). But as far as we know, no mutation of FLG has been found in IV patient. This result suggests that an unknown gene may underlie the pathology of IV disorder. In this study, we collected two affected Chinese IV families with highly similar histological features for linkage analysis, and localized the disease locus to chromosome 1q22.
Subjects and methods
Two three-generation families with isolated autosomal dominant IV were ascertained through initial identification of two probands from Sichuan Province in China. All family members received careful skin examination by an experienced dermatologist. All affected individuals had no evidence of progressive skin disorder. After informed consent was obtained, venous blood samples were collected from affected and unaffected family members. Genomic DNA was extracted from peripheral blood by use of the QIAmp DNA Blood kit (Qiagen, Germany).
In linkage studies, we chose microsatellite markers from the Genome Database. PCR amplifications were carried out according to the manufacturer's recommendation (LI-COR, USA) with a PTC-225 DNA Engine Tetrad (MJ Research, USA). PCR products were electrophoresed on 6% polyacrylamide-7 M urea gels with fluorescent-dye-labeled DNA markers using the LI-COR 4200L DNA sequencer (LI-COR). Following electrophoresis, data were collected and analyzed with Base Image 4.1 and Gene Image 3.12 software (LI-COR). Linkage ready pedigree files were prepared by the use of Gene Image software.
In linkage and haplotype analysis, autosomal dominant inheritance with 100% penetrance was assumed. The affected allele frequency was taken as 0.0001. Marker allele frequencies were set to be uniformly distributed. Two-point linkage analysis was performed using the MLINK and ILINK programs version 5.10 from the Linkage package (Lathrop et al. 1984). Multipoint analysis was conducted by FASTLINK Version 4.1P (Cottingham et al. 1993; Schaffer et al. 1994). Haplotype construction was performed by use of the program SimWalk2 version 2.52 with the aid of Cyrillic Version 2.02 software (Sobel and Lange 1996).
Results and discussion
We first examined the FLG gene in both IV families by sequencing the coding region and the promoter region. We did not detect any mutation in this gene. To identify the disease locus of IV, we performed a genome-wide search using linkage analysis. After yielding a weak supportive Lod score from DNA mark D1S3249 (Zmax=1.81, at θ=0.00), we typed all individuals with more high-density markers that spanned the genetic interval between D1S1665 and D1S1678. Two-point Lod scores for markers covering the candidate interval are summarized in Table 1. The maximum two-point Lod score of 2.47 is obtained at D1S1653 (θ=0.00).
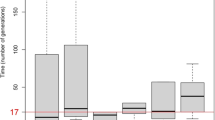
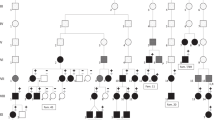
To determine the smallest interval containing the IV locus, recombination events among the family members were analyzed by haplotype reconstruction. The cosegregating segment in which recombination was not detected was flanked by the markers D1S1653 and D1S2675 (Fig. 1a). The locus was mapped to a 7-cM (4-Mb) region on 1q22. A multipoint maximum Lod score of 3.62 was obtained at the genetic position of D1S1167 (Fig. 1b).
Ichthyosis vulgaris pedigree. Haplotypes are shown for all available members with marker names at the left of each generation. Black bars represent disease-carrying haplotypes. The initial probands were indicated by arrows. b Multipoint linkage analysis for ichthyosis vulgaris and microsatellite DNA markers
The epidermal differentiation complex (EDC) comprises a remarkable cluster of genes that play an important role in terminal differentiation of the human epidermis (Marenholz et al. 1996). This cluster localizes within a 2-Mb region on 1q21. However, there is no overlap between the EDC and the IV locus reported here. This result rules out the candidate genes within EDC. Considering the short physical distance between the EDC and D1S1653 (only 4 Mb), we suggest that a novel gene on 1q22 might underlie the pathology of IV disorder.
References
Compton JG, DiGiovanna JJ, Johnston KA, Fleckman P, Bale SJ (2002) Mapping of the associated phenotype of an absent granular layer in ichthyosis vulgaris to the epidermal differentiation complex on chromosome 1. Exp Dermatol 11:518–526
Cottingham RW Jr, Idury RM, Schaffer AA (1993) Faster sequential genetic linkage computations. Am J Hum Genet 53:252–263
Hernandez-Martin A, Gonzalez-Sarmiento R, De Unamuno P (1999) X-linked ichthyosis: an update. Br J Dermatol 141:617–627
Lathrop GM, Lalouel JM, Julier C, Ott J (1984) Strategies for multilocus linkage analysis in humans. Proc Natl Acad Sci USA 81:3443–3446
Lei G, Zhang Y, Hu Y (1992) Investigation on the prevalence of Ichthyosis in Sichuan Province. Chin J Dermatol (in Chinese) 25:105–106
Marenholz I, Volz A, Ziegler A, Davies A, Ragoussis I, Korge BP, Mischke D (1996) Genetic analysis of the epidermal differentiation complex (EDC) on human chromosome 1q21: chromosomal orientation, new markers, and a 6-Mb YAC contig. Genomics 37:295–302
Nirunsuksiri W, Presland RB, Brumbaugh SG, Dale BA, Fleckman P (1995) Decreased profilaggrin expression in ichthyosis vulgaris is a result of selectively impaired posttranscriptional control. J Biol Chem 270:871–876
Okano M, Kitano Y, Yoshikawa K, Nakamura T, Matsuzawa Y, Yuasa T (1988) X-linked ichthyosis and ichthyosis vulgaris: comparison of their clinical features based on biochemical analysis. Br J Dermatol 119:777–783
Presland RB, Boggess D, Lewis SP, Hull C, Fleckman P, Sundberg JP (2000) Loss of normal profilaggrin and filaggrin in flaky tail (ft/ft) mice: an animal model for the filaggrin-deficient skin disease ichthyosis vulgaris. J Invest Dermatol 115:1072–1081
Schaffer AA, Gupta SK, Shriram K, Cottingham RW Jr (1994) Avoiding recomputation in linkage analysis. Hum Hered 44:225–237
Sobel E, Lange K (1996) Descent graphs in pedigree analysis: applications to haplotyping, location scores, and marker-sharing statistics. Am J Hum Genet 58:1323–1337
Wells RS, Kerr CB (1966) Clinical features of autosomal dominant and sex-linked ichthyosis in an English population. Br Med J 1:947–950
Acknowledgements
We are grateful to all of the individuals described here for their contribution to this study, and thank Jesse Pang for critical reading of this manuscript. This work was supported by the National High Technology "863" Programs of China and the National Science Fund for Distinguished Young Scholars.
Author information
Authors and Affiliations
Corresponding author
Additional information
W. Zhong and B. Cui contributed equally to this work
Rights and permissions
About this article
Cite this article
Zhong, W., Cui, B., Zhang, Y. et al. Linkage analysis suggests a locus of ichthyosis vulgaris on 1q22. J Hum Genet 48, 390–392 (2003). https://doi.org/10.1007/s10038-003-0043-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10038-003-0043-1
Keywords
This article is cited by
-
Exacerbation of ichthyosis vulgaris phenotype by co-inheritance of STS and FLG mutations in a Chinese family with ichthyosis: a case report
BMC Medical Genetics (2018)
-
Identification of a Genetic Locus for Ichthyosis Vulgaris on Chromosome 10q22.3–q24.2
Journal of Investigative Dermatology (2008)
-
Prevalent and Rare Mutations in the Gene Encoding Filaggrin Cause Ichthyosis Vulgaris and Predispose Individuals to Atopic Dermatitis
Journal of Investigative Dermatology (2006)
-
Loss-of-function mutations in the gene encoding filaggrin cause ichthyosis vulgaris
Nature Genetics (2006)