Abstract
A new antibiotic, designated sagamilactam, was isolated from the cultured broth of Actinomadura sp. K13-0306 by Diaion HP-20, silica gel and octadecylsilane column chromatography followed by purification by HPLC. The chemical structure of the new compound was elucidated by spectroscopic analyses, including NMR and MS. The structure of sagamilactam proved to be a new compound consisting of a polyunsaturated and polyoxygenated 34-membered macrocyclic lactam containing diene, triene and tetraene conjugated olefins and a decalin moiety. Sagamilactam showed antitrypanosomal activity with an IC50 value of 0.25±0.11 μM.
Similar content being viewed by others
Introduction
In our group, bioassay-guided fractionation of microbial cultured extracts has been employed to search for new bioactive compounds, and many useful compounds such as avermectin1 and lactacystin2 were discovered from the extracts. However, dereplication of already known compounds and their potential analogs is a vital part of the discovery process of new microbial natural products. Recently, it has become easy to acquire the UV and MS spectra of many single components of microbial cultured broths in combination with HPLC. We have focused on identifying new compounds from cultured broths by employing an approach based on the physicochemical properties of compounds using LC-UV/vis diode array detection, LC/MS and color reaction, collectively designated as physicochemical screening. For discovery of new compounds from actinomycetes, therefore, we have employed chromatographic and spectrometric methods in combination with natural product databases such as the Dictionary of Natural Products (http://dnp.chemnetbase.com/), as well as in-house databases. This physicochemical screening has identified new compounds such as spoxazomicins,3 trehangelins4 and mangromicins5, 6, 7 among the metabolites of actinomycete strains.
UV/vis and MS spectra-guided fractionation of the culture broth of actinomycete strain K13-0306, together with integrated LC/diode array detection and LC/MS techniques, has led to discovery of a new compound (designated sagamilactam (1)) consisting of a 34-membered macrocyclic lactam containing polyenes and a decalin moiety. The actinomycete strain K13-0306 was isolated from a soil sample collected in Kanagawa Prefecture, Japan. This paper deals with taxonomic studies of the producing strain, and isolation, physicochemical properties, structural elucidation and biological activity of the new compound.
Results
Taxonomy of the producing strain, Actinomadura sp. K13-0306
Strain K13-0306 was isolated from a soil sample collected in Kanagawa Prefecture, Japan. The strain grew well on ISP media 2, 3 and 6, but weakly on ISP media 4, 5 and 7. The colony color was yellow to grayish red. Extremely sparse white aerial mycelia were produced on ISP media 3 and 4, forming spiral spore chains (Figure 1). No soluble pigment was produced. Cell-wall peptidoglycans of strain K13-0306 contained meso-diaminopimelic acid as diamino acids. Glucose, galactose, madurose and ribose were detected from whole-cell hydrolysates. The predominant menaquinones of this strain were MK-9(H6) and MK-9(H8). Phosphatidylglycerol and phosphatidylinositol were detected but phosphatidylethanolamine and phosphatidylcolinewere not detected.8 The virtually complete 16S rRNA gene sequence (1432 bp) of strain K13-0306 was analyzed using the EzTaxon-e database (http://eztaxon-e.ezbiocloud.net/) and showed 98.4% similarity with Actinomadura bangladeshensis 3-46-b(3)T (AB331652).9 On the basis of the morphology, chemical composition and 16S rRNA gene sequence analyses, strain K13-0306 is identified as belonging to the genus Actinomadura.
Physicochemical screening
Twenty-three well-growing strains were selected from among 76 actinomycete isolates and cultured in soybean meal medium for LC/DAD and LC/MS analyses as described in the Methods. The Actinomadura sp. K13-0306 strain was selected based on the following criteria. (1) Many compounds (many peaks by LC/diode array detection analyses) were detected in the cultured broth of this isolate. (2) The product variety was strongly dependent on the producing culture medium. (3) The UV/vis spectrum of one product had a characteristic feature. Although many flavonoids and two decatromicins were detected from the cultured broth of strain K13-0306, the compound of interest showed unique and characteristic physicochemical properties including maximum absorption wavelengths of 270, 279 and 331 nm, along with an exact MS value of m/z 570.3529 (Figure 2). This compound (1) was deduced to have a novel structure based on database searches for these UV and MS spectra.
Isolation of sagamilactam (1)
The procedure for isolation of 1 is summarized in Scheme 1. A 10-l volume of cultured broth was centrifuged for 15min at 3000 r.p.m. The supernatant was passed over a Diaion HP-20 column (100-mm i.d. × 250 mm; Mitsubishi Chemical, Tokyo, Japan). The water-soluble materials were removed by washing with water, and the fraction containing 1 was eluted with 100% MeOH. As 1 was also contained in mycelia, the mycelial cake was extracted with MeOH and then filtered. Both samples from supernatant and mycelial cake were combined and concentrated in vacuo. The residue material was extracted with EtOAc. The EtOAc extracts were concentrated in vacuo under reduced pressure to yield 3.2 g of crude material. This material was applied to a silica gel FL100D column (60-mm i.d. × 200 mm; Fuji Silysia Chemical, Aichi, Japan) and eluted using a stepwise gradient of CHCl3-MeOH (100:0, 100:1, 100:2, 10:1, 1:1 and 0:100 (v/v)); the eluate was collected as six fractions. The eluate fraction (10:1 fraction) that contained 1 was concentrated to yield 311 mg. This material was applied to an octadecylsilane (ODS) column (40-mm i.d. × 150 mm; Senshu Scientific, Tokyo, Japan) previously equilibrated with water. After washing with water and 70% MeOH, the fractions containing 1 were eluted with 80% MeOH and concentrated in vacuo. The eluate fraction (37.8 mg) was dissolved in a small amount of MeOH and purified by HPLC on a PEGASIL ODS SP100 column (10-mm i.d. × 250 mm; Senshu Scientific) with 60% MeOH at 4.7 ml min−1 and subsequently detected at UV 270 nm. The yield of 1 was 8.4 mg.
Physicochemical properties of sagamilactam (1)
The physicochemical properties of 1 are summarized in Table 1. The molecule is readily soluble in MeOH, EtOH and pyridine but not in H2O and CHCl3. Compound 1 showed absorption maxima at 270, 279 and 331 nm in UV spectrum. The broad IR absorption at 3400 cm−1 suggested the presence of hydroxyl groups.
Structure elucidation of sagamilactam (1)
Compound 1 was obtained as a white powder and was determined to have a molecular formula of C37H47O4N by HR-electrospray ionization (ESI)-MS [M+H]+ m/z 570.3572 (consistent with a calculated value of 570.3578 for C37H48O4N) and NMR spectral data. The 1D and 2D NMR spectra of 1 were obtained primarily in CD3OD.
The 1H and 13C NMR spectral data of 1 are listed in Table 2. The 1H NMR data indicated the presence of 8 sp3 methine protons containing 3 oxygenated and 1 nitrogenated methines, 21 olefinic protons, 4 methylenes, 1 primary methyl and 1 secondary methyl. The 13C NMR spectrum showed the resonances of 37 carbons that were classified into 22 olefinic carbons, 1 carbonyl carbon at δc 173.7, 3 oxygenated sp3 methine carbons, 4 sp3 methine carbons, 1 nitrogenated sp3 methine carbon, 4 sp3 methylene carbons and 2 methyl carbons by HSQC spectra.
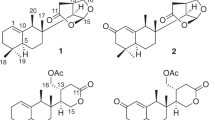
The 1H–1H COSY and TOCSY (total correlation spectroscopy) revealed the presence of four partial structures: (a) C-3/C-10, (b) C-13/C-29, (c) C-30/C-32 and (d) C-34/C-36, as shown in Figure 3a. Analysis of heteronuclear multiple bond correlation data confirmed the presence of a tetraene moiety, including partial structure a, based on correlations from H3-2-Me to C-1, C-2 and C-3; from H-3 to C-1, C-2-Me, C-4 and C-5; from H-4 to C-3 and C-5; from H-6 to C-8; from H-7 to C-6 and C-8; from H-8 to C-6 and C-10; from H-9 to C-7 and C-8; and from H-10 to C-9; a diene moiety, including partial structure b, based on correlations from H-19 to C-20; from H-20 to C-22; from H-21 to C-19 and C-23; from H-22 to C-24; and from H-23 to C-21 and C-24; a triene moiety, including partial structures b and c, based on correlations from H-26 to C-28; from H-27 to C-25, C-26, and C-29; from H-28 to C-26; from H-30 to C-28 and C-31; and from H-31 to C-30; a decalin moiety, including partial structure b, based on correlations from H-10 to C-11, C-12 and C-19; from H-11 to C-13; from H-12 to C-10 and C-18; from H2-14 to C-18; from H-15 to C-17; from H2-16 to C-15 and C-17; from H-18 to C-12; and from H-19 to C-10, C-11, C-13 and C-18; and a 1-propene moiety, including partial structure d, based on correlations from H-33 to C-34; from H-34 to C-33 and C-36-Me; from H-35 to C-33 and C-36-Me; and from H3-36-Me to C-34 and C-35.
The heteronuclear multiple bond correlations from H-8 to C-10, from H-10 to C-9, from H-11 to C-9, from H-19 to C-9 and C-20, from H-20 to C-10 and C-19 and from H-21 to C-19 confirmed a decalin ring linked between the C-9/C-10 and C-19/C-20 position; the correlations from H-22 to C-24, from H-23 to C-24 and C-25, from H-24 to C-25, from H-26 to C-24 and from H-27 to C-25 and C-26 confirmed a diene moiety was linked to a triene moiety through a methylene (C-24) and a hydroxyl carbon (C-25); and the correlations from H2-32 to C-34 and from H-33 to C-34 and C-35 confirmed a 1-propene moiety linked to the C-33 position. Finally, the planar structure of 1 as shown in Figure 3a was elucidated by 1H–13C and 1H–15N heteronuclear multiple bond correlations from H-33 to C-1 and from H2-32 and H-34 to NH-33, respectively.
The ROESY (rotating-frame overhauser effect spectroscopy) correlations were observed between H-9/H-18, H-10/H-20, H-13/H-17 and H-20 and H-19/H-21 in the decalin moiety. These results indicated that H-10, H-13 and H-17 are located on the same face of the decalin moiety. The double-bond geometries were used to determine the trans/cis form based on the coupling constants observed in double-quantum filtered COSY and ROESY (Supplementary Figures S4 and S9). The geometries of the double bonds in 1 were established from the ROESY correlations and the 1H–1H coupling constants. The ROESY coupling between H-21/H-23 and H-28/H-30 and their large coupling constants (15.0 Hz) established 20E, 22E and 28E geometries, and between H-26/H-27 (10.5 Hz) and H-30/H-31 (10.3 Hz) established 26Z and 30Z geometries. The ROESY coupling between 2-Me and H-4 and the trans-coupling constant (15.0 Hz) between H-2/H-3, H-4/H-5 and H-6/H-7 determined 2E, 4E and 6E, and the cis-coupling constant (10.3 Hz) between H-8/H-9 determined 8Z. The last double-bond geometry was determined as 34E based on the coupling constant of H-34/H-35 (18.0 Hz) (Figure 3b). The ROESY coupling between H-24a/H-26 and the coupling constants were observed between H-24a and H-25 (10.5 Hz), H-24b and H-25 (4.0 Hz) and H-25 and H-26 (10.5 Hz). These results reveal that H-25 methine is located on the opposite surface from H-24a and H-26. Overall, the structure of 1 proved to be a new polyunsaturated and polyoxygenated 34-membered macrocyclic lactam containing diene, triene and tetraene conjugated olefins along with a decalin moiety (Figure 4). However, the determination of absolute configuration of 1 will require further study.
Biological activities of sagamilactam (1)
Compound 1 was evaluated in an antiparasitic activity for a parasitic protozoan. As shown in Table 3, 1 showed more potent activity against Trypanosoma brucei brucei GUTat 3.1 strain than that of the clinically used antitrypanosomal drugs, suramin and eflornithine. The IC50 value of 1 was 0.14±0.06 μg ml−1 (0.25±0.11 μM).
Discussion
Many polyene macrocyclic lactam such as leinaistatin (DC107),10 hitachimycin11 and ML-449,12 which are 18- to 22-membered compounds, have been isolated as metabolites of Streptomyces sp. and Micromonospora sp. Members of the family Micromonosporaceae have also been reported to produce 24- and 26-membered macrocyclic lactam compounds such as sceliphrolactam,13 micromonosporin A14 and salinilactam.15 On the other hand, Actinomadura vulgaris and Actinomadura sp. SCC 1778 are known as producers of fluvirucin B116 and macrolactams,17 respectively, that are different types of 14-membered macrocyclic lactams that lack olefin moieties. Sagamilactam (1), produced by Actinomadura sp. K13-0306, is a new 34-membered macrocyclic lactam containing diene, triene and tetraene conjugated olefins and a decalin skeleton, suggesting the involvement of a possible Diels–Alder reaction in the biosynthesis of the decalin moiety. To our knowledge, 1 constitutes the largest known cyclic lactam produced by an actinomycete.
Macrocyclic lactams are less frequently reported as products of actinomycete strains than macrocyclic lactones, despite the structural similarities of the two classes. Nonetheless, lactam compounds, like macrolides, have been shown to exhibit a wide variety of biological activities. For instance, our group has previously identified hitachimycin in the culture broth of Streptomyces scabrisporus KM-4927T. Hitachimycin is a 19-membered macrocyclic lactam antibiotic with antiprotozoal properties, and also shows antibacterial, antifungal and antitumor activities.18
Recently, a novel biologically active polyene macrocyclic lactam, labosamide A and B, was isolated from the metabolites of a marine-derived Micromonospora sp. Labosamide A and B inhibited the growth inhibition of Trypanosoma brucei (IC50 <10 μM), the causative agent of human African trypanosomiasis.19 Compound 1 exhibited more potent growth inhibitory activity against T. brucei (IC50 0.25 μM) than those of labosamide A and B. Thus, macrocyclic lactams, including 1, may serve as starting points for the development of novel antitrypanosomal agents.
Methods
General experimental procedures
Silica gel and a Pegasil ODS SP100 column were purchased from Fuji Silysia Chemical and Senshu Scientific, respectively. All solvents for HPLC were purchased from Wako Pure Chemical (Osaka, Japan) or Kanto Chemical (Tokyo, Japan). NMR spectra were obtained using Varian XL-400 and INOVA 600 (Varian, Palo Alto, CA, USA) with 1H NMR at 400 or 600 MHz and 13C NMR at 100 or 150 MHz in CD3OD. The chemical shifts are expressed in ppm and are referenced to residual CHD2OD (3.31 ppm) in the 1H NMR spectra and CD3OD (49.0 ppm) in the 13C NMR spectra. LC-ESI-MS spectra were obtained using an AB Sciex QSTAR Hybrid LC/MS/MS Systems (AB Sciex, Framingham, MA, USA). IR spectra (KBr) were obtained using a Horiba FT-710 Fourier transform IR spectrometer (Horiba, Kyoto, Japan). UV spectra were obtained with a Hitachi U-2810 spectrophotometer (Hitachi, Tokyo, Japan). Optical rotation was measured on a JASCO model DIP-1000 polarimeter (Jasco, Tokyo, Japan).
Selection of strains and instrumental analyses of metabolites
A total of 76 rare, non-Streptomyces actinomycete strains were isolated from the roots and surrounding soil of Angelica keiskei growing at the medicinal plant botanical garden of Kitasato University, Sagamihara, Kanagawa Prefecture, Japan. A subset of 23 of these strains was selected as good growers. These strains were inoculated producing medium. These 23 strains were incubated on a reciprocating shaker (280 r.p.m.) for 6–8 days at 27 °C using 10- ml tubed (ϕ 20 × 250 mm) volumes of each of four different kinds of media. The media used were soybean meal medium (2.2% soluble starch, 2.2% glycerol, 1.2% meat extract, 2.0% peptone, 3.3% soybean meal and 2.2% CaCO3), starch medium (2.4% starch, 0.1% glucose, 0.5% peptone, 0.5% yeast extract, 0.3% meat extract and 0.4% CaCO3), Pharmamedia medium (0.5% glucose, 0.5% corn steep powder, 1.0% oatmeal, 1.0% Pharmamedia, 0.5% K2HPO4, 0.5% MgSO4·7H2O and 1 ml l−1 trace metals solution) and defatted wheat germ medium (2.0% soluble starch, 0.5% glycerol, 1.0% defatted wheat germ, 0.3% meat extract, 0.3% dry yeast and 0.3% CaCO3). Following culturing, each broth was combined with an equal volume of ethanol and then filtered. The filtrates were analyzed by LC/DAT and LC/MS. The resultant sample was injected into the electrospray ion source of a QSTAR Elite ESI quadruple time-of-flight (Q-TOF) MS instrument (AB Sciex) coupled to an Agilent 1200 series (Agilent Technologies, Santa Clara, CA, USA). Chromatographic separation was performed on an Inertsil ODS-4 column (3.0 × 250 mm, GL Sciences, Tokyo, Japan) at 40 °C. For gradient elution, solvent A was water with 0.1% formic acid, and solvent B was methanol with 0.1% formic acid. The gradient elution was programmed as follows: 0–30 min, 5–100% B. The flow rate was 0.5 ml min−1, the injection volume was 5 μl and UV detection was carried out by diode array detection. ESI-MS spectra were recorded for 30 min in the m/z region from 100 to 2000 Da with the following instrument parameters: ion spray voltage=5500 V, source gas=50 l min−1, curtain gas=30 l min−1, declustering potential= 50 V, focusing potential=250 V, temperature=450 °C and detector voltage=2300 V. LC/MS analysis was performed by HR-ESI-MS (R ⩾10 000, the tolerance for mass accuracy is 10 p.p.m.).
Taxonomic studies of strain K13-0306
Taxonomic studies of strain K13-0306 were carried out as described previously.4 The International Streptomyces Project media recommended by Shirling and Gottlieb20 and by Waksman21 were used to investigate cultural characteristics. Cultures were observed after incubation for 2 weeks at 27 °C. The morphological characteristics were observed by scanning electron microscope (JSM-5600; JEOL) after incubation on inorganic salts–starch agar for 3 weeks at 27 °C and fixation with 4% osmium tetroxide vapor. Isomers of diaminopimelic acid in whole-cell hydrolysates were determined by TLC, following the standard methods of Becker et al.22 and Hasegawa et al.23 Menaquinones were extracted and purified by the method of Collins et al.24 and then analyzed by LC/MS (JSM-T 100LP; JEOL) with a CAPCELL PAK C18 column (Shiseido, Tokyo, Japan) eluted with methanol/2-propanol (7:3). 16S rRNA gene sequence was amplified by PCR and sequenced on a 3130 Genetic Analyzer (Applied Biosystems, Carlsbad, CA, USA) using a BigDye Terminator v.3.1 cycle sequencing kit (Applied Biosystems) according to the manufacturer’s instructions.
Fermentation
Strain K13-0306 was grown and maintained on freeze broth consisting of 1.0% dextrose and 1.0% yeast extract (adjusted to pH 7.0 before sterilization). A loop of spores of strain K13-0306 was inoculated into 100 ml of the seed medium consisting of 1.0% dextrose and 1.0% yeast extract (adjusted to pH 7.0 before sterilization) in a 500-ml Erlenmeyer flask. The flask was incubated on a rotary shaker (210 r.p.m.) at 27 °C for 7 days. A 2-ml portion of the seed culture was transferred to each of 100 500-ml Erlenmeyer flasks, each containing 100 ml of SBM medium consisting of 3.3% soybean meal, and fermentation was carried out on a rotary shaker (210 r.p.m.) at 27 °C for 7 days.
Antitrypanosomal activity in vitro
The growth inhibitory effect of 1 and reference compounds against T. brucei brucei strain GUTat 3.1 were measured by the previously described method.24 In brief, GUTat 3.1 was cultured in Iscove’s modified Dulbecco’s media with various supplements and 10% heat-inactivated fetal bovine serum at 37 °C, under 5.0% CO2 and 95% air. Subsequently, 95 μl of the trypanosome suspension (2.0–2.5 × 104 trypanosomes per ml) were transferred into each well of a 96-well microtiter plate and 5.0 μl of a test compound solution (dissolved in 5.0% DMSO) was added to a given well. The plate was then incubated for 72 h at 37 °C (long incubation–low inoculation test). Subsequently, 10 μl of the fluorescent dye Alamar Blue was added to each well. After incubation for 3–6 h, the resulting solution was read at 528/20 nm excitation wavelengths and 590/30 nm emission wavelengths using an FLx800 fluorescence microplate reader (BioTek Instruments, Winooski, VT, USA). The IC50 values were determined using the fluorescent plate reader software (KC-4; BioTek Instruments). Successive subcultures were carried out in 24-well tissue culture plates under the same conditions.
Assessment of the cytotoxic effect of 1 and reference compounds against human fetal lung fibroblast MRC-5 cells was carried out as described previously.25

Purification of sagamilactam (1).
References
Burg, R. W. et al. Avermectins, new family of potent anthelmintic agents: producing organism and fermentation. Antimicrob. Agents Chemother. 15, 361–367 (1979).
Ōmura, S. et al. Lactacystin, a novel microbial metabolite, induces neuritogenesis of neuroblastoma cells. J. Antibiot. 44, 113–116 (1991).
Inahashi, Y. et al. Spoxazomicins A-C, novel antitrypanosomal alkaloids produced by an endophytic actinomycete, Streptosporangium oxazolinicum K07-0460T. J. Antibiot. 64, 303–307 (2011).
Nakashima, T. et al. Trehangelins A, B and C, novel photo-oxidative hemolysis inhibitors produced by an endophytic actinomycete, Polymorphospora rubra K07-0510. J. Antibiot. 66, 311–317 (2013).
Nakashima, T. et al. Mangromicins A and B: structure and antitrypanosomal activity of two new cyclopentadecane compounds from Lechevalieria aerocolonigenes K10-0216. J. Antibiot. 67, 253–260 (2014).
Nakashima, T., Kamiya, Y., Iwatsuki, M., Takahashi, Y. & Ōmura, S Mangromicins, six new anti-oxidative agents isolated from a culture broth of the actinomycete, Lechevalieria aerocolonigenes K10-0216. J. Antibiot. 67, 533–539 (2014).
Nakashima, T. et al. Mangromicin C, a new analog of mangromicin. J. Antibiot. 68, 220–222 (2015).
Lechevalier, M. P. et al. Chemotaxonomy of aerobic actinomycetes: phospholipid composition. Biochem. Syst. Ecol. 5, 249–260 (1977).
Ara, I. et al. Actinomadura bangladeshensis sp. nov. and Actinomadura chokoriensis sp. nov. Int. J. Syst. Evol. Microbiol. 58, 1653–1659 (2008).
Hara, M. et al. DC 107, a novel antitumor antibiotic produced by a Streptomyces sp. J. Antibiot. 42, 333–335 (1989).
Ōmura, S., Nakagawa, A., Shibata, K. & Sano, H. The structure of hitachimycin, a novel macrocyclic lactam involving beta-phenylalanine. Tetrahedron Lett. 23, 4713–4716 (1982).
Jørgensen, H. et al. Insights into the evolution of macrolactam biosynthesis through cloning and comparative analysis of the biosynthetic gene cluster for a novel macrocyclic lactam, ML-449. Appl. Environ. Microbiol. 76, 283–293 (2010).
Oh, D. C., Poulsen, M., Currie, C. R. & Clardy, J. Sceliphrolactam, a polyene macrocyclic lactam from a wasp-associated Streptomyces sp. Org. Lett. 13, 752–755 (2011).
Thawai, C. et al. Micromonosporin A, a novel 24-membered polyene lactam macrolide from Micromonospora sp. isolated from peat swamp forest. Chem. Biodivers. 1, 640–645 (2004).
Udwary, D. W. et al. Genome sequencing reveals complex secondary metabolome in the marine actinomycete Salinispora tropica. Proc. Natl Acad. Sci. USA 104, 10376–10381 (2007).
Lin, T. Y. et al. Sequence, cloning, and analysis of the fluvirucin B1 polyketide synthase from Actinomadura vulgaris ACS Synth. Biol 2, 635–642 (2013).
Cooper, R. et al. Macrolactams: two novel homologous series of compounds produced by Actinomadura sp. SCC 1778. J. Antibiot. 45, 633–638 (1992).
Shibata, K. et al. Chemical modification of hitachimycin. Synthesis, antibacterial, cytocidal and in vivo antitumor activities of hitachimycin derivatives. J. Antibiot. 41, 614–623 (1988).
Schulze, C. J. et al. Genome-directed lead discovery: biosynthesis, structure elucidation, and biological evaluation of two families of polyene macrolactams against Trypanosoma brucei. ACS Chem. Biol. 10, 2373–2381 (2015).
Shirling, E. B. & Gottlieb, D. Methods for characterization of Streptomyces species. Int. J. Syst. Bacteriol. 16, 313–340 (1966).
Waksman, S. A. (ed) The Actinomycetes Vol. 2, (Williams and Wilkins, Baltimore, (1961).
Becker, B., Lechevalier, M. P. & Lechevalier, H. A. Chemical composition of cell-wall preparation from strains of various form-genera of aerobic actinomycetes. Appl. Microbiol. 13, 236–243 (1965).
Hasegawa, T., Takizawa, M. & Tanida, S. A rapid analysis for chemical grouping of aerobic actinomycetes. J. Gen. Appl. Microbiol. 29, 319–322 (1983).
Collins, M. D., Goodfellow, M. & Minnikin, D. E. Distribution of menaquinones in actinomycetes and corynebacteria. J. Gen. Microbiol. 100, 221–230 (1977).
Otoguro, K. et al. Potent antimalarial activities of polyether antibiotic, X-206. J. Antibiot. 54, 658–663 (2001).
Acknowledgements
We are grateful to Dr Kenichiro Nagai, School of Pharmacy, Kitasato University, for determination of the MS. This study was supported by funds from the Institute for Fermentation (IFO), Osaka, Japan.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies the paper on The Journal of Antibiotics website
Supplementary information
Rights and permissions
About this article
Cite this article
Kimura, T., Iwatsuki, M., Asami, Y. et al. Anti-trypanosomal compound, sagamilactam, a new polyene macrocyclic lactam from Actinomadura sp. K13-0306. J Antibiot 69, 818–824 (2016). https://doi.org/10.1038/ja.2016.28
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ja.2016.28
This article is cited by
-
Genomic insight into a novel actinobacterium, Actinomadura rubrisoli sp. nov., reveals high potential for bioactive metabolites
Antonie van Leeuwenhoek (2021)
-
Hamuramicins A and B, 22-membered macrolides, produced by an endophytic actinomycete Allostreptomyces sp. K12-0794
The Journal of Antibiotics (2018)
-
Structure elucidation and in silico docking studies of a novel furopyrimidine antibiotics synthesized by endolithic bacterium Actinomadura sp. AL2
World Journal of Microbiology and Biotechnology (2017)