Abstract
To identify the mutations in multi- and extensively drug-resistant tuberculosis isolates and to evaluate the use of molecular markers of resistance, we analyzed 257 multi- and extensively drug-resistant isolates and 64 pan-sensitive isolates from 23 provinces in China. Seven loci associated with drug resistance, including rpoB for rifampin (RIF), katG, inhA and oxyR-ahpC for isoniazid (INH), gyrA and gyrB for ofloxacin (OFX), and rrs for kanmycin (KAN), were examined by DNA sequencing. Compared with the phenotypic data, the sensitivity and specificity for DNA sequencing were 91.1% and 98.4% for RIF, 80.2% and 98.4% for INH, 72.2% and 98.3% for OFX and 40% and 98.2% for KAN, respectively. The most common mutations found in RIF, INH, OFX and KAN resistance were Ser531Leu (48.2%) in rpoB, Ser315Thr (49.8%) in katG, C(−15)T (10.5%) in inhA, Asp94Gly (20.3%), Asp94Ala (12.7%) and Ala90Val (21.5%) in gyrA, and A1401G (40%) in rrs. This molecular information will be helpful to establish new molecular biology-based methods for diagnosing multi- and extensively drug-resistant tuberculosis in China.
Similar content being viewed by others
Introduction
Drug-resistant tuberculosis is a growing global problem and a major public health concern.1 China is one of the countries in which tuberculosis is widespread. A recent (2007) national survey of drug-resistant tuberculosis isolates in China indicated that ~5.7% of new cases of tuberculosis and 25.6% of previously treated cases were multidrug-resistant tuberculosis (MDR-TB) strains (strains resistant to at least rifampin (RIF) and isoniazid (INH)).2 Among the MDR-TB isolates, ~8% were extensively drug-resistant tuberculosis (XDR-TB),2 defined as tuberculosis strains resistant to any fluoroquinolone and to at least one of the three injectable drugs of capreomycin, amikacin and kanmycin (KAN), in addition to being MDR. Due to the long treatment phase, high treatment cost, unsatisfactory therapeutic efficacy and high mortality, the emergence of MDR-TB and XDR-TB has become a major threat to the control of tuberculosis.1
Abuse of antibiotics is one of the important causes of drug resistance. Accurate and rapid detection of drug susceptibility is crucial for the timely implementation of an effective therapeutic regimen to interrupt the transmission of MDR- and XDR-TB. However, traditional culture-based drug susceptibility testing is limited by the slow growth characteristics of Mycobacterium tuberculosis (M. tuberculosis). Previous reports showed that resistance to tuberculosis was usually associated with certain genetic mutations.3, 4 Furthermore, the characteristics of the mutations showed some diversity in different counties and regions. Hence, studying the characteristics of gene mutations among MDR- and XDR-TB isolates in China is imperative for the establishment of rapid and accurate diagnostics methods to be implemented in China.
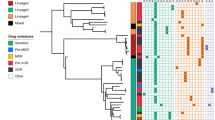
Although there are several reports on the mutated features of MDR-TB in local areas of China,3, 5, 6 information regarding the mutations among MDR-TB isolates remains unclear in many regions of China. This study is the first to include a large number of MDR- and XDR-TB isolates (230 MDR- and 27 XDR-TB isolates, and 64 pan-sensitive isolates) from 23 provinces of China, to determine the drug resistance mutations. In addition, seven drug resistance-associated loci, including katG, inhA and OxyR-ahpC intergenic region, rpoB, gyrA, gyrB and rrs, were analyzed in our research.
Materials and Methods
Ethical approval
The study was approved by the Ethics Committee of the National Institute for Communicable Disease Control and Prevention, Chinese Center for Disease Control and Prevention. The patients with tuberculosis included in this study were given a Subject information sheet and they all gave written informed consent to participate in the study.
M. tuberculosis isolates
A total of 257 M. tuberculosis identified as MDR-TB isolates were recovered from patients with pulmonary tuberculosis in 23 different provinces in China (Fujian, Guangdong, Guangxi, Hunan, Hubei, Jiangxi, Chongqing, Sichuan, Anhui, Zhejiang, Gansu, Guizhou, Xizang, Henan, Hebei, Beijing, Shandong, Shanxi, Shaanxi, Ningxia, Inner Mongolia, Jilin and Heilongjiang). For every province, 8–15 MDR- and XDR-TB strains were selected. Sixty-four isolates identified as pan-susceptible strains were selected as negative controls using a random number table matched by region and isolation time. The H37Rv (ATCC 27294) strain was used as the reference.
Drug susceptibility testing
All of the isolates were cultured on Lowenstein-Jensen solid medium. The drug susceptibility testing was routinely determined via the 1% indirect proportions method utilizing Lowenstein–Jensen solid medium supplemented individually with the following drugs: INH (0.2 μg ml−1), RIF (40.0 μg ml−1), OFX (2.0 μg ml−1) and KAN (30.0 μg ml−1).7
DNA isolation, PCR amplification and sequencing
Genomic DNA was extracted from M. tuberculosis strains using the CTAB (cetyltrimethylammonium bromide)-NaCl method.8 The DNA was dissolved in pure water and stored at −20 °C. The following seven loci were amplified by PCR: rpoB, katG, inhA, oxyR-ahpC, gyrA, gyrB and rrs. The primer sequences and amplicon positions are presented in Supplementary Table 1. The target fragments were amplified in a 30 μl reaction volume, containing 15 μl of 2xTaq master mix, 0.5 μl (25 μm) of forward and reverse primer, 1 μl of genomic DNA (~100 ng) and 13 μl of distilled H2O. The amplification parameters consisted of an initial denaturation at 95 °C for 5 min, followed by 35 cycles of denaturation at 95 °C for 40 s, annealing at 64 °C for 40 s, elongation at 72 °C for 1 min and a final elongation at 72 °C for 7 min. The amplified products underwent electrophoresis in a 1.5% agarose gel and were sent for sequencing. All sequence data were compared with the reference M. tuberculosis H37Rv strain (GenBank accession number NC_000962) using BioEdit version 7.05.3 (Tom Hall, North Carolina State University, Raleigh, NC, USA).
Resolution of discrepancy
The tests were repeated with both methods in the event of a discrepancy between the DNA sequencing and drug susceptibility testing results. If the repeated results were inconsistent with the initial data, a third round of detection was performed and the concordance result was used.
Statistical analysis
All data were analyzed with SPSS 16.0 software. Percentages and confidence intervals (CIs) were calculated as appropriate. The χ2-test was used for comparisons. Differences were considered statistically significant at a P-value of <0.05.
Results
RIF and rpoB
A 543-bp fragment containing the RIF-resistant determining region (RRDR) of 81 bp in rpoB was examined in this study. In total, among the 257 MDR-TB isolates, 234 (91.1%) isolates contained at least one mutation (including insertions and deletions), whereas the remaining 23 isolates (8.9%) lacked such mutations (Supplementary Table 2). Notably, 99.6% (233 isolates) of the isolates carried mutations within the rpoB RRDR region, and 1 isolate (0.4%) contained a mutation not in the rpoB RRDR. The most frequent mutations were in codons 531, 526 and 516 (Table 1), which accounted for 82.1% of the RIF-resistant isolates (Supplementary Table 2). Of 64 pan-susceptible isolates, only 1 double mutation (Ile572Phe/Ser574Leu) was identified outside the rpoB RRDR. The detection of mutations in rpoB exhibited a sensitivity of 91.1% and a specificity of 98.4% compared with conventional phenotypic drug susceptibility testing (Table 2).
INH and inhA, katG and oxyR-ahpC
The majority of INH resistance-associated mutations were in katG, the inhA promoter and the intergenic region oxyR-ahpC. Our results demonstrated that 80.2% of INH-resistant isolates harbored a mutation within at least one of these three regions. One-hundred and thirty isolates harbored mutations only in katG, 20 isolates harbored mutations only in inhA and 32 isolates harbored mutations only in oxyR-ahpC (Supplementary Table 3). Twenty-four isolates harbored mutations in two regions. The most common mutations were katG315 (140 isolates), especially for Ser315Thr (128 isolates; Table 1) and inhA C(−15)T (27 isolates). Two novel mutations, the C(−34) deletion and C(−34)T, were identified in inhA. Another novel mutation, C(−26)T in inhA, was also identified in one pan-susceptible isolate. Hence, the combination of mutations within katG, inhA and OxyR-ahpC intergenic regions could detect INH resistance with a sensitivity of 80.2% and a specificity of 98.4%, better than that obtained using the data for any single gene mutation (Table 2).
OFX and gyrA and gyrB
Among the 79 isolates resistant to OFX, a total of 53 isolates (67.1%) carried mutations in gyrA, including codons 94, 90 and 91 (Supplementary Table 4). Codon 94 was the most prevalent mutation site; others included 16 isolates with Asp94Gly, 9 isolates with Asp94Ala, 3 isolates with Asp94Asn, 2 isolates with Asp94Tyr, 2 isolates with Asp94His and 1 isolate with Asp94Phe (Supplementary Table 4). One isolate with Asp94Ala also had Ser91Pro. Ala90Val was the next most prevalent mutation, with 17 isolates presenting this mutant type. All tuberculosis clinical isolates possessed a substitution of Ser95Thr in gyrA. No mutation other than Ser95Thr was observed in OFX-susceptible isolates. Six OFX-resistant strains carried mutations within gyrB, including Arg446His (n=3), Asp461Asn (n=2) and Gly512Arg (n=1). Double mutations in gyrA and gyrB were detected in two isolates. Four OFX-susceptible isolates displayed mutations in gyrB: one isolate with Arg446His, one isolate with Gln503His and two isolates with Gly512Arg. Sequence analyses of gyrA and gyrB had a sensitivity and specificity of 72.2% and 98.3%, respectively (Table 2).
KAN and rrs
Sixteen of 40 KAN-resistant isolates contained the transition A1401G in rrs. Moreover, we also found five KAN-susceptible MDR-TB isolates carrying A1449G, C1459G, G1473T or T1491A (Supplementary Table 5). No mutation was observed in rrs among 64 pan-susceptible isolates. The rrs mutation displayed a susceptibility of 40% and a specificity of 98.2% for the detection of KAN resistance among the strains analyzed.
Discussion
Rapid and reliable diagnostic methods are urgently required to control the rise in MDR-TB. Determining the type and frequency of specific mutations associated with drug resistance is crucial to develop rapid and accurate molecular diagnostic methods. Therefore, we investigated the type and frequency of common drug resistance-conferring mutations that occurred in a large number of MDR- and XDR-TB isolates from multiple Chinese provinces.
Mutations in rpoB are the main cause of RIF resistance. Our results showed a sensitivity of 91.1% for phenotypic RIF resistance among MDR-TB, in accordance with the data from some regions of China and Argentina (P>0.05).5, 9, 10 The highest frequency of RIF mutations is in the rpoB RRDR region, particularly in codons 531, 526 and 516.3, 4, 5, 10 Our study verified these findings: 82.1% of MDR-TB isolates had a mutation in one of these three codons.
By contrast, resistance to INH is more complex, and mutations within katG and the promoter of inhA account for the majority of INH-resistant isolates. Mutations in the intergenic region of oxyR-ahpC are another important cause of INH resistance. In our study, the sensitivity was 58.4% for katG, 14.4% for inhA, and 67.7% for the two genes together. This sensitivity is lower than that reported from America and Africa (P<0.05).4, 10, 11 We also determined that 32 (12.5%) isolates had mutations only in oxyR-ahpC. When the mutations in oxyR-ahpC were considered in association with katG and inhA, the sensitivity of detecting INH resistance increased to 80.2%, suggesting that mutations in oxyR-ahpC were significant for detecting INH resistance among MDR-TB isolates in China. Two novel mutations in inhA, the C(−34) deletion and C(−34)T, were identified in the present study. Further research is required to determine their roles. Furthermore, 51 MDR-TB isolates possessed no mutation within the three genes, which suggests that there may be alternative mechanisms of INH resistance, such as an active drug efflux pump.12, 13
Fluoroquinolone is an important second-line anti-tuberculosis drug for therapy in patients with MDR-TB. It could inhibit the function of DNA gyrase, which is encoded by gyrA and gyrB. Thus, mutations in gyrA and gyrB give rise to fluoroquinolone resistance. The gyrA QRDR region, particularly in codons 94, 90 and 91, has been considered as the primary source of mutations.14, 15, 16 In our study, 67.1% of OFX-resistant isolates carried at least one mutation in the three codons, lower than the surveys in the United States and Argentina (P<0.05),4, 10 but similar to other reports from China (P>0.05).16, 17 This suggests that there are regional variations in gyrA mutations. In addition, Ser95Thr was observed in all 321 clinical isolates. This mutation is not associated with OFX resistance, but is a naturally occurring polymorphism. For mutations in gyrB, only 7.6% of the mutations occurred in OFX-resistant isolates. Arg446His in gyrB occurred in three OFX-resistant isolates and one OFX-sensitive isolate, which was thought to confer a low-level resistance.16, 18 In addition, gyrB Gly512Arg and gyrA Ala90Val together occurred in one OFX-resistant isolate, but gyrB Gly512Arg was also the only mutation in two OFX-sensitive isolates, suggesting that Gly512Arg alone does not confer OFX resistance. In addition, 22 OFX-resistant isolates (27.8%) were wild-type for either gyrA or gyrB. Hence, some other causes, such as an active drug efflux pump, permeability reduction of the bacterial cell wall19, 20 or hetero-resistance,21 may contribute to the development of OFX resistance.
KAN is an important second-line injectable drug for MDR-TB treatment. Resistance to KAN mostly develops as a result of alterations in rrs coding for 16S ribosomal RNA. A1401G in rrs has been identified as the primary cause of resistance to KAN, which correlates with high-level resistance.22 In our study, 40% of the KAN-resistant MDR-TB possessed the A1401G mutation, consistent with the data from the United States (P>0.05),4 but lower than the findings from some regions (P<0.05).5, 10 This difference is probably attributable to geographical differences. Clearly, there are other mechanisms such as a mutation in eis,23 and other unknown mechanisms that have a role in KAN resistance. Four mutations, A1449G, C1459G, G1473T and T1491A, were identified in KAN-sensitive strains, which were not reported to confer KAN resistance in previous publications.
Compared with the phenotypic results, the specificity for detecting INH, RIF, KAN and OFX resistance by DNA sequencing in the present study was 98.4% (95% CI, 97.0–99.8%), 98.4% (95% CI, 97.0–99.8%), 98.3% (95% CI, 96.9–99.7%) and 98.2% (95% CI, 96.7–99.7%), respectively, in agreement with the range of 93.6 to 100.0% in the literature (P>0.05).3, 4, 5, 10
Notably, some drug resistance could not be explained in the present study. Thus, whole-genome sequencing might be required to search for additional loci conferring drug resistance and to promote the performance of the sequencing-based assay.
In general, the establishment of rapid molecular methods requires the verification of molecular data with respect to phenotypic results. Here, we accomplished a survey of seven loci known to harbor mutations conferring resistance to first- and second-line anti-tuberculosis drugs among a large number of MDR/XDR-TB cases in China. The results indicated that DNA sequencing could detect 91.1% of RIF, 80.2% of INH, 72.2% of OFX and 40.0% of KAN resistance among MDR-TB isolates in China. Moreover, we also identified the predominant mutations among MDR-TB isolates in China as rpoB531, katG315, inhA-15, gyrA94 and -90 and rrs1401. This information is useful to develop rapid diagnostic tests, such as reverse dot blot hybridization assays,24, 25 gene chips26, 27 and multiplex allele-specific PCR,28, 29, 30, 31 for faster detection of MDR/XDR-TB, more appropriate anti-tuberculosis treatment regimens and reduced transmission of drug-resistant TB.
Accession codes
References
Gandhi, N. R. et al. Multidrug-resistant and extensively drug-resistant tuberculosis: a threat to global control of tuberculosis. Lancet 375, 1830–1843 (2010).
Zhao, Y. et al. National survey of drug-resistant tuberculosis in China. N. Engl. J. Med. 366, 2161–2170 (2012).
Luo, T. et al. Selection of mutations to detect multidrug-resistant Mycobacterium tuberculosis strains in Shanghai, China. Antimicrob. Agents Chemother. 54, 1075–1081 (2010).
Campbell, P. J. et al. Molecular detection of mutations associated with first- and second-line drug resistance compared with conventional drug susceptibility testing of Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 55, 2032–2041 (2011).
Yuan, X. et al. Molecular characterization of multidrug- and extensively drug-resistant Mycobacterium tuberculosis strains in Jiangxi, China. J. Clin. Microbiol. 50, 2404–2413 (2012).
Zhao, L. L. et al. Molecular characterization of multidrug-resistant Mycobacterium tuberculosis isolates from China. Antimicrob. Agents Chemother. 58, 1997–2005 (2014).
WHO Policy Guidance on TB Drug Susceptibility Testing (DST) of Second-Line Drugs. WHO/HTM/TB/2008392. WHO: Geneva, Switzerland, (2008).
Somerville, W., Thibert, L., Schwartzman, K. & Behr, M. A. Extraction of Mycobacterium tuberculosis DNA: a question of containment. J. Clin. Microbiol. 43, 2996–2997 (2005).
Yue, J. et al. Mutations in the rpoB gene of multidrug-resistant Mycobacterium tuberculosis isolates from China. J. Clin. Microbiol. 41, 2209–2212 (2003).
Engstrom, A., Morcillo, N., Imperiale, B., Hoffner, S. E. & Jureen, P. Detection of first- and second-line drug resistance in Mycobacterium tuberculosis clinical isolates by pyrosequencing. J. Clin. Microbiol. 50, 2026–2033 (2012).
Feuerriegel, S. et al. Sequence analysis for detection of first-line drug resistance in Mycobacterium tuberculosis strains from a high-incidence setting. BMC Microbiol. 12, 90 (2012).
Machado, D. et al. Contribution of efflux to the emergence of isoniazid and multidrug resistance in Mycobacterium tuberculosis. PLoS ONE 7, e34538 (2012).
Rodrigues, L., Machado, D., Couto, I., Amaral, L. & Viveiros, M. Contribution of efflux activity to isoniazid resistance in the Mycobacterium tuberculosis complex. Infect. Genet. Evol. 12, 695–700 (2012).
Ali, A. et al. Characterization of mutations conferring extensive drug resistance to Mycobacterium tuberculosis isolates in Pakistan. Antimicrob. Agents Chemother. 55, 5654–5659 (2011).
Hu, Y. et al. Prevalence and genetic characterization of second-line drug-resistant and extensively drug-resistant Mycobacterium tuberculosis in Rural China. Antimicrob. Agents Chemother. 57, 3857–3863 (2013).
Zhang, Z., Lu, J., Wang, Y., Pang, Y. & Zhao, Y. Prevalence and molecular characterization of fluoroquinolone-resistant Mycobacterium tuberculosis isolates in China. Antimicrob. Agents Chemother. 58, 364–369 (2014).
Zhao, L. L. et al. Prevalence and molecular characteristics of drug-resistant Mycobacterium tuberculosis in Hunan, China. Antimicrob. Agents Chemother. 58, 3475–3480 (2014).
Nebenzahl-Guimaraes, H., Jacobson, K. R., Farhat, M. R. & Murray, M. B. Systematic review of allelic exchange experiments aimed at identifying mutations that confer drug resistance in Mycobacterium tuberculosis. J. Antimicrob. Chemother. 69, 331–342 (2014).
Singh, M. et al. Effect of efflux pump inhibitors on drug susceptibility of ofloxacin resistant Mycobacterium tuberculosis isolates. Indian J. Med. Res. 133, 535–540 (2011).
Wolfe, L. M., Mahaffey, S. B., Kruh, N. A. & Dobos, K. M. Proteomic definition of the cell wall of Mycobacterium tuberculosis. J. Proteome. Res. 9, 5816–5826 (2010).
Eilertson, B. et al. High proportion of heteroresistance in gyrA and gyrB in fluoroquinolone-resistant Mycobacterium tuberculosis clinical isolates. Antimicrob. Agents Chemother. 58, 3270–3275 (2014).
Du, Q. et al. Mycobacterium tuberculosis rrs A1401G mutation correlates with high-level resistance to kanamycin, amikacin, and capreomycin in clinical isolates from mainland China. Diagn. Microbiol. Infect. Dis. 77, 138–142 (2013).
Zaunbrecher, M. A., Sikes, R. D. Jr, Metchock, B., Shinnick, T. M. & Posey, J. E. Overexpression of the chromosomally encoded aminoglycoside acetyltransferase eis confers kanamycin resistance in Mycobacterium tuberculosis. Proc. Natl Acad. Sci. USA 106, 20004–20009 (2009).
Suresh, N. et al. Rapid detection of rifampicin-resistant Mycobacterium tuberculosis by in-house, reverse line blot assay. Diagn. Microbiol. Infect. Dis. 56, 133–140 (2006).
Mokrousov, I. et al. Multicenter evaluation of reverse line blot assay for detection of drug resistance in Mycobacterium tuberculosis clinical isolates. J. Microbiol. Methods 57, 323–335 (2004).
Zhang, Z. et al. Rapid and accurate detection of RMP- and INH- resistant Mycobacterium tuberculosis in spinal tuberculosis specimens by CapitalBio DNA microarray: a prospective validation study. BMC Infect. Dis. 12, 303 (2012).
Guo, Y. et al. Rapid, accurate determination of multidrug resistance in M. tuberculosis isolates and sputum using a biochip system. Int. J. Tuberculos. Lung Dis. 13, 914–920 (2009).
Evans, J. & Segal, H. Novel multiplex allele-specific PCR assays for the detection of resistance to second-line drugs in Mycobacterium tuberculosis. J. Antimicrob. Chemother. 65, 897–900 (2010).
Vadwai, V., Shetty, A. & Rodrigues, C. Multiplex allele specific PCR for rapid detection of extensively drug resistant tuberculosis. Tuberculosis (Edinb) 92, 236–242 (2012).
Chia, B. S. et al. Use of multiplex allele-specific polymerase chain reaction (MAS-PCR) to detect multidrug-resistant tuberculosis in Panama. PLoS ONE 7, e40456 (2012).
Zhao, L. L. et al. Multiplex allele-specific PCR combined with PCR-RFLP analysis for rapid detection of gyrA gene fluoroquinolone resistance mutations in Mycobacterium tuberculosis. J. Microbiol. Methods 88, 175–178 (2012).
Acknowledgements
We thank the staff of Fujian, Guangdong, Guangxi, Hunan, Hubei, Jiangxi, Chongqing, Sichuan, Anhui, Zhejiang, Gansu, Guizhou, Xizang, Henan, Hebei, Beijing, Shandong, Shanxi, Shaanxi, Ningxia, Inner Mongolia, Jilin and Heilongjiang for supplying strains. This study was supported by the projects of National Natural Science Foundation of China (Grant No. 81201348) and the National Key Program of Mega Infectious Diseases (Grant No. 2013ZX10003002-001). The funder had no role in the study design, data collection and analysis, decision to publish or preparation of the manuscript.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies the paper on The Journal of Antibiotics website
Supplementary information
Rights and permissions
About this article
Cite this article
Chen, Y., Zhao, B., Liu, Hc. et al. Prevalence of mutations conferring resistance among multi- and extensively drug-resistant Mycobacterium tuberculosis isolates in China. J Antibiot 69, 149–152 (2016). https://doi.org/10.1038/ja.2015.106
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ja.2015.106