Abstract
The methods for typing and epidemiological study for especially antibiotic-resistant bacteria has been issued but there are the debates regarding which method is best for this purpose. The purpose of this study is to investigate and apply a comparatively new technology, phage-open-reading frame typing (POT) and repetitive-sequence-based PCR (rep-PCR) using DiversiLab system and compare for the discrimination of major methicillin-resistant Staphylococcus aureus (MRSA) lineages in epidemiological surveillance. We analyzed 47 representative MRSA stains isolated in Kobe University Hospital between January and December 2009. We performed MRSA typing using the POT kit and rep-PCR using the DiversiLab system. POT method classified all the MRSA strains into 35 clusters, whereas rep-PCR method typed all the MRSA strains in 10 kinds of clusters with a definition of 95% similarity. The discriminatory power and congruence between the methods were compared using the Simpson’s index of diversity, adjusted Rand’s and Wallace’s coefficients. Our statistical analyses showed that the POT (POT 1-2-3 and POT 2-3) revealed a higher discriminatory power in the Simpson’s index of diversity (SID; 0.969, range 0.939–1.000 and 0.967, range 0.935–0.998, respectively) for MRSA isolates than the rep-PCR (0.821 (0.767–0.876)). The adjusted Rand’s and Wallace’s coefficients did not show higher concordance among the methods. In conclusion, we demonstrated that the POT can perform accurate and reliable epidemiological surveillance studies for analyzing the genetic relatedness of MRSA strains.
Similar content being viewed by others
Introduction
Methicillin-resistant Staphylococcus aureus (MRSA), a representative antibiotic-resistant bacterium, is an increasingly important cause of mortality.1, 2 A rapid epidemiological surveillance system for screening for MRSA is necessary for preventing MRSA infections and their spread. The genetic relatedness of MRSA strains is an essential information for infection control and epidemiological studies. Several molecular typing methods including multilocus sequence typing, spa typing, repetitive-sequence-based PCR (rep-PCR) and pulsed-field gel electrophoresis (PFGE), have already been in use.3, 4, 5, 6 In particular, PFGE, a well-known highly discriminative molecular typing method for MRSA,3, 4 is limited by labor-intensive analyses and ambiguous data interpretation. A rapid epidemiological surveillance system for screening for MRSA is necessary for preventing MRSA infections. Rep-PCR for the rapid typing of MRSA has been reported as functionally improved and provided useful information within 8 h for the data of epidemiological surveillance.7, 8 A phage-open-reading frame typing (POT) method, which is a rapid, multiplex PCR-based method, amplifying the phage-open-reading frames of phage genomes, has been recently reported the methodology for the target including small genomic islets and Staphylococcal cassette chromosome mec elements of S. aureus by Suzuki et al.9 It has become a popular tool for epidemiological surveillance of community-and hospital-associated MRSA in Japan6, 10, and POT kit was shown as an excellent tool with high discriminatory power for the rapid detection of genotyping of skin and soft tissue infection-associated MRSA.6 Specially, POT methods for discrimination have three kinds of scores (POT1, POT2 and POT3) and their single use or combination use may have more potential for this purpose than single use only.
In this study, we directly compared the POT method including their single use or combination use of POT1, POT2 and POT3 scores with the rep-PCR method for epidemiologic analysis in a setting with multiple endemic MRSA strain types with the potential to cause epidemics.
Materials and Methods
MRSA strains
Forty-seven isolates collected from several kinds of materials including urine or blood from patients with infection by MRSA were obtained in Kobe University Hospital between January and December 2009 as described previous study.11 MRSAs were diagnosed by MRSA selective agar (Becton, Dickinson and Company, Franklin Lakes, NJ, USA).
POT
Bacterial strains were cultured on tryptic soy agar plates for 18 h and were then cultured in lysogeny broth medium for 8 h. DNA extraction was performed with a QIAamp DNA Mini kit (Qiagen, Tokyo, Japan). We performed the POT method using the Cica Geneus Staph POT Kit (Kanto Chemical, Tokyo, Japan) according to the manufacturer’s protocol as described previous study.11 The POT method was presented by being categorized as three POT scores (POT 1, POT 2 and POT 3) by a combination of the presence or absence of 22 genetic elements6, 9 using the aforementioned POT kit according to the manufacture’s protocol. In brief, (1) overnight culture at 37 °C of MRSA strain, (2) DNA extraction, (3) 4% agarose electrophoresis, (4) Exchange to final POT typing (POT 1, POT 2 and POT 3). For the improvement of recognition potential, we employed three POT scores (POT 1, POT 2 and POT 3). For the calculation of POT scores, we employed the open-reading frames for detection as follows: reaction mixture 1 consists of femA (target region: S. aureus positive control), POT 1-1 (mecA), POT 1-2 (mec gene complex class B), POT1-3 (SCCmec typeΠ a specific), POT 2-1 (Tn554), POT 2-2 to 7 (prophages) and POT 2–8 (Genomic Island), and reaction mixture 2 consists of femA (S. aureus positive control), POT 1–4 (cassette chromosome recombinase A2), POT 1–5 (genetic background), POT 1–6 (genetic background), POT 1–7 (mec gene complex class A), POT 3-1 to7 (prophages). Then, final POT 1 was totaled by the amount of POT 1-1 to 7 × POT coefficient, which was assigned in each POT scores such as 1, 2, 4, 8, 16, 32, 64, 128, 256, and final POT 2 was the amounts of POT 2-1 to 8, and final POT 3 was the amounts of POT 3-1 to 7.
Rep-PCR
Bacterial strains were subcultured in 5% sheep blood agar medium with 48 h of incubation. Bacterial DNA was extracted using the UltraClean microbial DNA isolation kit (bioMérieux, Marcy l’Etoile, France). Rep-PCR amplification for each species was performed with the designated DiversiLab Staphylococcus kit (bioMérieux). The amplified rep-PCR products were separated and detected on a microfluidic chip by the Agilent 2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA, USA) and analyzed by DiversiLab analysis software (version 3.4; bioMérieux) to construct a genealogical tree from the calculation with Pearson’s product-moment correlation coefficient and unweighted pair-group method using arithmetic averages for average linkages for cluster analysis. The independent three assays were performed for the observation of reproducibility.
Data analysis
POT and rep-PCR were compared for their discriminatory power using Simpson's index of diversity (SID) (with 95% confidence intervals).12, 13 Specially, POT score diagnoses were performed by a single use of POT score or the combination use of POT scores such as POT1-2-3 or POT 2-3. The congruence between the typing methods was calculated using the adjusted Rand’s and Wallace’s coefficients.14, 15 The SID should be at least in the order of 0.90–0.95 for a typing system to be considered robust.4 The adjusted Rand’s coefficient is used to measure the overall agreement between two typing methods adjusted for chance match.15 The Wallace’s coefficients are an estimate of how much additional information was provided by data from another typing method.14 The adjusted Rand’s coefficient indicated that a lower score (<0.5) was the less congruent between the two methods, while a higher score (>0.8) was as congruent between them. A high value of Wallace’s coefficient (>0.6) indicates that partitions defined by a given method could have been predicted from the results of another method, suggesting that the use of both methodologies is redundant.14 All calculations were done using EpiCompare software (Ridom Bioinformatics, Münster, Germany, http://www.ridom.de).
Results
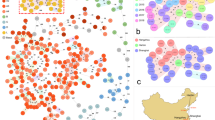
In POT method, all 47 MRSA strains, dividing into POT1, POT2-3 or POT 1-2-3 were classified to 9, 33 or 35 different types, respectively (Table 1 and Figure 1). In the whole group of clinical MRSA isolates, 10 kinds of MRSA clusters were detected in rep-PCR (Table 1 and Figure 1). The reproducibility study demonstrated tight clustering (>95%) of isolates, supporting the good reliability of the rep-PCR typing results. The SID for POT1-2-3 was the largest for MRSA (0.969 (range: 0.939–1.000)). The SID for POT 2-3 (0.967 (0.935–0.998)) also demonstrated a high value, while those for POT1 (0.708 (0.607–0.809)) and rep-PCR (0.821 (0.767–0.876)) did not. This means that POT1-2-3 and POT2-3 appear to have a higher discriminatory power than POT1 or rep-PCR. POT (POT 1-2-3, POT 1 and POT 2-3) and rep-PCR had low congruence with the lower adjusted Rand’s and Wallace’s coefficients by all combinations (crude: 0.625–0.797 and adjusted: −0.022 to −0.019 for Rand’s coefficient and 0.021–0.269 for Wallace’s coefficients), suggesting that taken together, statistical data of SID in our tested four methods revealed that POT1-2-3 and POT2-3 may have significant discriminatory power for MRSA strains compared with rep-PCR and the concordance between POT 1-2-3, POT 1, and POT 2-3, and rep-PCR was not significant as their typing methods.
(a) Similarity dendrogram of repetitive-sequence-based PCR (rep-PCR patterns), (b) rep-PCR patterns and (c) Phage-open-reading frame typing (POT) patterns shown by POT 1-2-3 scores of digested chromosomal DNA from methicillin-resistant Staphylococcus aureus (MRSA) strains isolated from multiple specimens were shown. The letters in the PCR column is the strain No. Regarding the No. 40 strain, we had no reproducible data of POT and changed to bar from 0 0 0 in the places where POT scores are written. NC, negative control; PC, positive control; SL, singleton.
Discussion
This study compared POT method and rep-PCR method for the epidemiological investigation of clinical MRSA strains originating from different regions and materials. To our knowledge, this is the first report of direct comparison between POT and rep-PCR, although other reports have been compared5, 6 in Enterobacter spp., MRSA, E. coli, Acinetobacter spp. or Stenotrophomonas maltophilia for this purpose. The ability to perform rapid, accurate outbreak analyses to detect the origin and spread of epidemic hospital infections is a critical need for keeping patients’ health.16, 17 The methodological aspects of this epidemiological study allowed us to compare several modalities and exploit the modality with better outcomes and lower cost.18, 19 In this study, our epidemiological investigation used clinical MRSA-related infectious isolates over a 1 year period, rather than isolates from one particular MRSA outbreak as seen in most studies. Therefore our targeted bacteria were expected to have diverse MRSA strain backgrounds and different time courses of isolation. Typing MRSA strains gathered in this way may include several or multiple kinds of strains and represents a technical challenge and so it may be important to see which modality for investigation of MRSA typing in this case of POT or rep-PCR.
Several groups have reported epidemiological analyses of MRSA infections for detecting bacterial relatedness. Clarridge et al.20 distinguished MRSA strains from nares and wound isolates by several diagnostic modalities such as rep-PCR, Spectra Cell Raman analysis, PFGE, an antibiotic susceptibility profile and whole-genome optical mapping (OpGen, Gaithersburg, MD, USA) was performed on selected isolates in a patient population and reported the variations based on the interpretation method and discriminatory power of strain typing.19, 21 Overall categorization of strain types depends on the methodology and interpretation criteria and not all analyses are equivalent.17, 22 This was reflected in our data as well, which showed some inconsistency between POT and rep-PCR results.
In our study, the adjusted Rand’s and Wallace’s coefficients offered very low congruence between POT and rep-PCR; this may be partly because the targets of POT1 are small genomic islets and SCCmec and targets of POT2 and POT3 are phage.9, 23 Moreover, we have reported that the MRSA strains with higher MICs to Linezolid showed that this strain was classified as SCCmec type IIa by POT analysis from its band pattern.11 Maeda et al.6 showed the effectiveness of the POT method for SCCmec typing in skin and soft tissue infection (SSTI)-MRSA in comparison with PFGE, spa typing and multilocus sequence typing, with a simple and rapid platform producing results in as little as 6 h without nucleotide sequencing. They also showed that combined diagnoses by POT scores (POT1-2-3 and POT2-3) had higher discriminatory power but the concordance between POT1-POT2-POT3 and other genotyping methods was not excellent and our results were supported by their findings.
There is another opinion that rep-PCR was inadequate as a screening method for MRSA, because the coefficients of rep-PCR and spa typing or PFGE in MRSA were lower than that of other bacteria.5 The rep-PCR can be used for other clinically important microbes, such as Acinetobacter spp., Pseudomonas aeruginosa, Clostridium difficile or extended-spectrum beta-lactamase-producing Klebsiella spp.17, 24, 25 and this feature (with the shorter time course and simpler procedure and applicability to other microbes) should be useful in many institutions, not only clinical microbiology laboratories but also hospitals, which deal with many patients with infectious diseases. On the other hand, PFGE needs 2 to 4 days to get the final results depending on the specific protocol but rep-PCR is able to get results within 1 day.19 They also concluded that though PFGE showed greater discriminatory power, rep-PCR is a fast and simple diagnostic method of molecular fingerprinting that can be used as an initial evaluation tool for investigating S. aureus isolates.
These features of the methods for typing antibiotic-resistant strains such as MRSA suggest that, taken together, POT may have a benefit to rep-PCR in the viewpoint of coefficient value with conventional methods such as spa typing22 and to PFGE in the viewpoint of the required time duration for results for the purpose of not only typing but epidemiological methods.6
We would like to emphasize the study limitations. The congruence of the results from two or more typing methods varies according to the size of the sample taken.15 Therefore, our data may differ from analyses using additional MRSA strains. Next, the number of MRSA strains investigated was not enough for definitive conclusion. We lack the comparisons with more conventional modalities such as PFGEs or spa typing. Third, this study describes the antibiotic-resistant bacteria mainly focusing on a kind of a new methodological aspect by POT and epidemiological point of views, suggesting that it might be lack of the research for the mode of action of antibiotics or mechanism of resistance to antibiotics. However, in the near future, we have a project to connect this study with the history of antibiotic dosing and the kinds of antibiotics such as cephalosporines or carbapenems for further expansion of this study. These limitations will be overcome by our future study.
In conclusion, our study demonstrated that the POT method was able to obtain accurate epidemiological information from a genetic relatedness analysis of MRSA-related infection strains to produce reliable epidemiological surveillance studies. This study will be strengthened by further examinations and comparisons employing a wider range of diagnostic modalities
References
Shurland, S., Zhan, M., Bradham, D. D. & Roghmann, M. C. Comparison of mortality risk associated with bacteremia due to methicillin-resistant and methicillin-susceptible Staphylococcus aureus. Infect. Control Hosp. Epidemiol. 28, 273–279 (2007).
Wolkewitz, M., Frank, U., Philips, G., Schumacher, M. & Davey, P. Mortality associated with in-hospital bacteraemia caused by Staphylococcus aureus: a multistate analysis with follow-up beyond hospital discharge. J. Antimicrob. Chemother. 66, 381–386 (2011).
Murchan, S. et al. Harmonization of pulsed-field gel electrophoresis protocols for epidemiological typing of strains of methicillin-resistant Staphylococcus aureus: a single approach developed by consensus in 10 European laboratories and its application for tracing the spread of related strains. J. Clin. Microbiol. 41, 1574–1585 (2003).
van Belkum, A. et al. Guidelines for the validation and application of typing methods for use in bacterial epidemiology. Clin. Microbiol. Infect. 13, S1–S46 (2007).
Fluit, A. C. et al. Evaluation of the DiversiLab system for detection of hospital outbreaks of infections by different bacterial species. J. Clin. Microbiol. 48, 3979–3989 (2010).
Maeda, T. et al. Genotyping of skin and soft tissue infection (SSTI)-associated methicillin-resistant Staphylococcus aureus (MRSA) strains among outpatients in a teaching hospital in Japan: application of a phage-open reading frame typing (POT) kit. J. Infect. Chemother. 18, 906–914 (2012).
Shutt, C. K., Pounder, J. I., Page, S. R., Schaecher, B. J. & Woods, G. L. Clinical evaluation of the DiversiLab microbial typing system using repetitive-sequence-based PCR for characterization of Staphylococcus aureus strains. J. Clin. Microbiol. 43, 1187–1192 (2005).
Tenover, F. C. et al. Comparison of typing results obtained for methicillin-resistant Staphylococcus aureus isolates with the DiversiLab system and pulsed-field gel electrophoresis. J. Clin. Microbiol. 47, 2452–2457 (2009).
Suzuki, M., Tawada, Y., Kato, M., Hayakawa, H. & Minagawa, H. Development of a rapid strain differentiation method for methicillin-resistant Staphylococcus aureus isolated in Japan by detecting phage-derived open-reading frames. J. Appl. Microbiol. 101, 938–947 (2006).
Moriyama, H. et al. Usefulness of phage ORF typing, a rapid genotyping method as a molecular Rand’s epidemiological method for detecting methicillin resistant Staphylococcus aureus. Kansenshogaku. Zasshi 86, 115–120 (2012).
Abe, Y., Shigemura, K., Yoshida, H., Fujisawa, M. & Arakawa, S. Risk factors for anti-MRSA drug resistance. Int. J. Antimicrob. Agents 40, 423–426 (2012).
Hunter, P. R. & Gaston, M. A. Numerical index of the discriminatory ability of typing systems: an application of Simpson’s index of diversity. J. Clin. Microbiol. 26, 2465–2466 (1988).
Grundmann, H., Hori, S. & Tanner, G. Determining confidence intervals when measuring genetic diversity and the discriminatory abilities of typing methods for microorganisms. J. Clin. Microbiol. 39, 4190–4192 (2001).
Carriço, J. A. et al. Illustration of a common framework for relating multiple typing methods by application to macrolide-resistant Streptococcus pyogenes. J. Clin. Microbiol. 44, 2524–2532 (2006).
Pinto, F. R., Melo-Cristino, J. & Ramirez, M. A confidence interval for the wallace coefficient of concordance and its application to microbial typing methods. PLoS ONE 3, e3696 (2008).
Bertin, M. L. et al. Outbreak of methicillin-resistant Staphylococcus aureus colonization and infection in a neonatal intensive care unit epidemiologically linked to a healthcare worker with chronic otitis. Infect. Control Hosp. Epidemiol. 27, 581–585 (2006).
Grisold, A. J. et al. Use of automated repetitive-sequence-based PCR for rapid laboratory confirmation of nosocomial outbreaks. J. Infect. 60, 44–51 (2010).
Olive, D. M. & Bean, P. Principles and applications of methods for DNA-Based typing of microbial organism. J. Clin. Microbiol. 37, 1661–1669 (1999).
Ross, T. L., Merz, W. G., Farkosh, M. & Carroll, K. C. Comparison of an automated repetitive sequence-based PCR microbial typing system to pulsed-field gel electrophoresis for analysis of outbreaks of methicillin-resistant Staphylococcus aureus. J. Clin. Microbiol. 43, 5642–5647 (2005).
Clarridge, J. E. 3rd, Harrington, A. T., Roberts, M. C., Soge, O. O. & Maquelin, K. Impact of strain typing methods on assessment of relationship between paired nares and wound isolates of methicillin-resistant Staphylococcus aureus. J. Clin.Microbiol. 51, 224–231 (2013).
Salmenlinna, S. et al. Molecular epidemiology of methicillin-resistant Staphylococcus aureus in Finland. Eur. J. Clin. Microbiol. Infect. Dis. 19, 101–107 (2000).
Babouee, B., Frei, R., Schultheiss, E., Widmer, A. F. & Goldenberger, D. Comparison of the DiversiLab repetitive element PCR system with spa typing and pulsed-field gel electrophoresis for clonal characterization of methicillin-resistant Staphylococcus aureus. J. Clin. Microbiol. 49, 1549–1555 (2011).
Suzuki, M., Matsumoto, M., Takahashi, M., Hayakawa, Y. & Minagawa, H. Identification of the clonal complexes of Staphylococcus aureus strains by determination of the conservation patterns of small genomic islets. J. Appl. Microbiol. 107, 1367–1374 (2009).
Brolund, A. et al. The DiversiLab system versus pulsed-field gel electrophoresis: characterisation of extended spectrum β-lactamase producing Escherichia coli and Klebsiella pneumoniae. J. Microbiol. Methods 83, 224–230 (2010).
Pasanen, T. et al. Comparison of repetitive extragenic palindromic sequence-based PCR with PCR ribotyping and pulsed-field gel electrophoresis in studying the clonality of Clostridium difficile. Clin. Microbiol. Infect. 17, 166–175 (2011).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Osawa, K., Shigemura, K., Jikimoto, T. et al. Comparison between phage-open-reading frame typing and automated repetitive-sequence-based PCR for typing MRSA isolates. J Antibiot 67, 565–569 (2014). https://doi.org/10.1038/ja.2014.41
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ja.2014.41