Abstract
A novel endophytic actinomycete, designated strain KLBMP 1483T, was isolated from the stem of the coastal plant Dendranthema indicum (Linn.) Des Moul collected from Nantong, in East China. Phylogenetic analysis showed that strain KLBMP 1483T was affiliated with the genus Glycomyces within the family Glycomycetaceae and shared the highest 16S rRNA gene sequence similarities with the type strains of Glycomyces arizonensis NRRL B-16153T (96.7%) and Glycomyces tenuis IFO 15904T (96.2%), and lower similarities (94.1–95.1%) to the other members of the genus Glycomyces, which distinguished KLBMP 1483T from representatives of the genus Glycomyces. The whole-cell hydrolysates contained meso-diaminopimelic acid, glucose, xylose and galactose. The polar lipids were diphosphatidylglycerol, phosphatidylglycerol, phosphatidylinositol, phosphatidylinositol mannosides, two unknown aminophospholipids, two phosphoglycolipids, two unknown phospholipids and one unknown lipid. MK-10(H4) was the predominant menaquinone. The major fatty acids were iso-C15:0, anteiso-C15:0, iso-C16:0, iso-C16:1 G and anteiso-C17:0. On the basis of the phenotypic and genotypic characteristics presented in this study, strain KLBMP 1483T represents a novel species, for which the name Glycomyces phytohabitans sp. nov. is proposed. The type strain is KLBMP 1483T (NBRC 109116T=DSM 45766T).
Similar content being viewed by others
Introduction
Actinomycetes are widely distributed in different habitats and are well-known producers of an enormous variety of secondary metabolites, including antibiotics, antitumor agents, enzyme and immunosuppressive agents.1 More recently, endophytic actinomycetes have attracted significant interest for their enormous biodiversity and capacity to produce a vast array of secondary metabolites.2, 3, 4, 5 During a study on the culturable diversity and bioactivities of endophytic actinomycetes from coastal halophytes, an aerobic actinomycete strain, KLBMP 1483T, was isolated from the stem of Dendranthema indicum (Linn.) Des Moul collected from Nantong, Jiangsu Province, East China. In this study, identification and classification of the isolate are reported by a polyphasic taxonomic study, with the proposal of the name Glycomyces phytohabitans sp. nov.
The genus Glycomyces representing Gram-positive actinobacteria belongs to class Actinobacteria, suborder Glycomycineae and family Glycomycetaceae.6 It was first reported by Labeda et al.7 in 1985 when the type strains of Glycomyces harbinensis NRRL 15337T and Glycomyces rutgersensis NRRLB-16106T were, respectively, isolated from soil samples from Harbin, People’s Republic of China and Rutgers University, NJ, USA during the course of isolation of actinomycete strains for antibiotic screening. Its description was later emended by Labeda and Kroppenstedt.8 Members of the genus are characterized chemotaxonomically by type II cell walls (meso-diaminopimelic acid and glycine are present) and type PI phospholipid pattern with significant amounts of phosphatidylinositol mannosides. The menaquinones predominantly contain 10, 11 and/or 12 isoprene units, but the degree of saturation varies within each species. In addition, the predominant fatty acids present in the members of this genus mostly consist of iso C15:0, anteiso C15:0, iso C16:0 and anteiso C17:0.8 At present, the genus Glycomyces consists of 11 species with validly published names: Glycomyces algeriensis, Glycomyces arizonensis, Glycomyces harbinensis, Glycomyces lechevalierae, Glycomyces rutgersensis, Glycomyces tenuis, Glycomyces sambucus, Glycomyces mayteni, Glycomyces endophyticus, Glycomyces scopariae and Glycomyces halotolerans (http://www.bacterio.net/g/glycomyces.html).7, 8, 9, 10, 11, 12, 13
Materials and methods
Strains and cultural conditions
Plant samples were collected from the coastal region of Nantong, Jiangsu Province, East China and used as the source for the isolation of endophytes. The samples were first surface sterilized using previously described five-step procedures.14 After that, the samples were aseptically crumbled into smaller fragments using a commercial blender and spread onto cellulose agar medium (per liter: cellulose 2.5 g, KNO3 1.0 g, MgSO4 0.2 g, K2HPO4 0.2 g, CaCl2 0.5 g, FeSO4 0.01 g, NaCl 30 g, 0.5 g amino-acid mixture and agar 15.0 g, pH 7.2). Strain KLBMP 1483T was maintained on ISP 4 agar15 containing 3% (w/v) NaCl at 4 °C for short-term preservation and as a glycerol suspension (20%, w/v in distilled water) at −80 °C for long-term preservation.
Morphological, physiological and biochemical characterization
Growth on various culture media was investigated by using yeast extract-malt extract agar (ISP 2), oatmeal agar (ISP 3), inorganic salts-starch agar (ISP 4), glycerol-asparagine agar (ISP 5),15 as well as potato-dextrose (PDA), Czapek’s and nutrient agars,16 for 21 days at 28 °C. The colony and mycelia color was determined with the ISCC–NBS color charts.17 Cell morphology was observed with light microscopy (SA3300-PL) and scanning electron microscopy (Hitachi, Tokyo, Japan; S-3400N) and cells grown for 28 days at 28 °C. Growth at different temperatures (4, 10, 15, 20, 28, 37, 45, 50 and 55 °C) was tested on ISP 2 agar containing 3% (w/v) NaCl. The ability of strain KLBMP 1483T to grow at different salt concentrations (1–15% NaCl, w/v, at intervals of 1%) was examined by growing the novel strain on ISP 2 basal medium. The ability to grow at different pH values (4.0 to 10.0, adjusted with 1.0 M HCl or 1.0 M NaOH, at intervals of 1 pH unit) was examined in ISP 2 broth at 28 °C. Carbon and nitrogen source utilization tests were assessed according to Kurup and Schmitt18 and Williams et al.19 Other phenotypic characteristics were carried out according to Gordon et al.20 All the media used were added with 3% (w/v) NaCl.
16S rRNA gene sequencing and phylogenetic analyses
Isolation of chromosomal DNA, PCR amplification and direct sequencing of the PCR products of isolate KLBMP 1483T were carried out as described by Qin et al.21 The identification of phylogenetic neighbors was initially carried out by using the BLAST program. Calculation of pairwise 16S rRNA gene sequence identities was achieved using the EzTaxon-e database.22 The phylogenetic relationship between the isolate and closely related strains was investigated using the neighbor-joining,23 maximum-parsimony24 and maximum-likelihood25 algorithms. Phylogenetic analysis was performed using MEGA version 5.26 Tree stability was assessed by comparison with other trees constructed with the maximum-parsimony and maximum-likelihood methods. The topology of the phylogenetic tree was evaluated by using the bootstrap resampling method of Felsenstein27 with 1000 replicates.
Chemotaxonomy
Chemotaxonomic characteristics of strain KLBMP 1483T were determined by using biomass obtained from cultures grown in ISP 4 broth for 7 days at 28 °C with shaking. Whole-cell sugars and isomers of diaminopimelic acid in whole-cell hydrolysates were prepared and analyzed by thin-layer chromatography.28 Purified cell wall was obtained by using the method of Kawamoto et al.,29 and the amino-acid composition of hydrolyzed cell walls was determined by thin-layer chromatography. The acyl type of the muramic acid in the cell wall was determined by the method of Uchida and Aida.30
Menaquinones were extracted and purified as described by Collins et al.31 and analyzed by HPLC.32 Analysis of polar lipids by thin-layer chromatography was performed as described by Minnikin et al.33 Cellular fatty acids were extracted from cells cultivated in ISP 2 broth at 28 °C for 7 days. Fatty acids were extracted, methylated and analyzed by using the Sherlock Microbial Identification System (MIDI) according to the manufacturer’s instructions. Fatty acid methyl esters were analyzed by gas chromatography using the Microbial Identification software package (MIDI, Sherlock Version 6.1; database, TSBA6; gas chromatograph, model 6890 N, Agilent Technologies, Santa Clara, CA, USA). The G+C content of the DNA was determined by using the method of Mesbah et al.34
Nucleotide sequence accession number
The 16S rRNA gene sequence of strain KLBMP 1483T determined in this study has been deposited in GenBank under the accession number JQ819256.
Results and Discussion
An almost complete 16S rRNA gene sequence (1436 nt) was obtained for the strain KLBMP 1483T. Comparative 16S rRNA gene sequence analysis showed that the strain was phylogenetically affiliated with species of Glycomyces, the highest similarities being found with the sequences of G. arizonensis NRRL B-16153T (96.7% 16S rRNA gene sequence similarity) and G. tenuis IFO 15904T (96.2% 16S rRNA gene sequence similarity). Levels of 16S rRNA gene sequence similarities of strain KLBMP 1483T with the other recognized species of the genus Glycomyces ranged from 94.1 to 95.1%. Similarities between the 16S rRNA gene sequences of KLBMP 1483T and all the Glycomyces species were observed to be less than 97%. It is generally accepted that organisms displaying 16S rRNA sequence similarity values of 97% or less belong to different species.35 Thus, the strain KLBMP 1483T represents a distinct species of the genus Glycomyces. It is evident from the phylogenetic tree (Figure 1) based on the neighbor-joining method that the strain KLBMP 1483T clusters with the nearest neighbors G. arizonensis NRRL B-16153T and G. tenuis IFO 15904T in a separate branch with high bootstrap support. This topology was also confirmed in the maximum-likelihood and maximum-parsimony trees.
Phylogenetic tree based on 16S rRNA gene sequence analysis, constructed using the neighbor-joining method showing the inter-relationship of strain KLBMP 1483T and type strains of related Glycomyces species. Numbers at branching points refer to bootstrap values of 1000 resamplings. The sequence of Bifidobacterium gallicum JCM 8224T (D86189) was used as outgroup. Asterisks indicate the clades that were conserved when maximum-parsimony and maximum-likelihood methods were used to construct phylogenetic trees. Bar, 0.01 substitutions per nucleotide position.
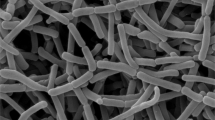
Morphological observation of the 4-week-old culture of strain KLBMP 1483T revealed that white aerial mycelia were produced on some of the tested media (ISP 3, ISP 4, NA and Czapek’s agar). The substrate mycelium varies from white to gray/deep gray. Black diffusible pigments were produced on ISP 3 and ISP 4 media agar. Strain KLBMP 1483T showed good growth on all the media tested except moderate growth on ISP 5 agar and poor growth on ISP 3 media. Chains of square-ended conidia were produced on aerial mycelia (Figure 2). Growth of the strain KLBMP 1483T occurred in the pH range 6.0–9.0 and 0–7% NaCl (w/v), with optimum growth at pH 7.0 and 3% NaCl (w/v). The temperature range for growth was 10–37 °C, with the optimum temperature being 28 °C. The detailed physiological features are indicated in Table 1 and in the species description. Several test results were obtained that enable the differentiation of strain KLBMP 1483T from its most closely related two Glycomyces species (Table 1).
The cell wall of strain KLBMP 1483T contained meso-diaminopimelic acid and glycine. Whole-cell hydrolysates contained glucose, xylose and galactose. The strain KLBMP 1483T was found to contain N-glycolylmuramic acid. The polar lipids detected in strain KLBMP 1483T were diphosphatidylglycerol, phosphatidylglycerol, phosphatidylinositol, phosphatidylinositol mannosides, two unknown aminophospholipids, two phosphoglycolipids, two unknown phospholipids and one unknown lipid (Supplementary Figure S1). The menaquinones determined in the strain KLBMP 1483T were MK-10 (4%), MK-10(H4) (71%), MK-11 (8%), MK-11(H2) (11%) and MK-11(H4) (6%). The predominant menaquinone components of strain KLBMP 1483T were similar to G. arizonensis NRRL B-16153T but different from the strain G. tenuis IFO 15904T (Table 1). Major fatty acids (>10%) detected in the strain KLBMP 1483T were iso-C15:0 (21.8%), anteiso-C15:0 (14.1%), iso-C16:1 G (11.4%), iso-C16:0 (16.3%) and anteiso-C17:0 (10.2%). Table 2 summarizes the cellular fatty acid profiles of strain KLBMP 1483T and its closest phylogenetic relative G. arizonensis NRRL B-16153T. The fatty acid profiles of strain KLBMP 1483T were similar to those members of the genus Glycomyces but differed from G. arizonensis NRRL B-16153T based on the presence or proportions of C18:1 ω9c, iso-C15:0, anteiso-C15:0, iso-C16:1 G and iso-C16:0. The DNA G+C content was 71.6 mol%, which is in accordance with its placement in the proposed genus too.
The morphological and chemotaxonomic properties of strain KLBMP 1483T were consistent with those members of the genus Glycomyces. The 16S rRNA gene sequence similarities between strain KLBMP 1483T and related Glycomyces species were low (<97%). Furthermore, the strain KLBMP 1483T can be distinguished from related phylogenetic closest species based on cultural and physiological and chemotaxonomic characteristics, as shown in Tables 1 and 2. Thus, on the basis of the phenotypic and genotypic characteristics, the strain KLBMP 1483T represents a novel species within the genus Glycomyces, for which the name Glycomyces phytohabitans sp. nov. is proposed.
Description of Glycomyces phytohabitans sp. nov
G. phytohabitans (Phy.to.ha'bi.tans. Gr. n. phyton, plant; L. part. adj. habitans, inhabiting; N. L. part. adj. used as a masc. n. phytohabitans, plant inhabiting, isolated from a plant).
Cells are aerobic and Gram positive. Aerial mycelia are present and vegetative mycelia have branched hyphae. Chains of square-ended conidia were produced on aerial mycelia. Black soluble pigment produced on some media. Temperature and pH ranges for growth are 10–37 °C and pH 6.0–9.0, with optimal at 28 °C and pH 7.0. The NaCl concentration range for growth is 0–7%, with optimal growth occurring at 3%. It is positive for nitrate reduction and milk peptonization and coagulation. Urease is positive. Casein, chitin, starch, Tween 20, Tween 40 and Tween 80 are hydrolyzed, but negative for gelatin liquefaction, cellulose hydrolysis and H2S production. D-Arabinose, D-cellobiose, D-fructose, D-galactose, D-glucose, lactose, maltose, D-mannose, rhamnose, D-ribose, trehalose and xylose are utilized as sole carbon sources, but not D-raffinose. Uses L-arginine, L-cysteine, L-proline and L-histidine as sole nitrogen sources, but negative for assimilation of L-alanine, L-asparagine, L-glycine, L-serine, L-tyrosine and L-lysine. Whole-cell hydrolysates contain meso-diaminopimelic acids, glycine, glucose, xylose and galactose. Menaquinones present include MK-10, MK-10(H4), MK-11, MK-11(H2) and MK-11(H4). The polar lipid profile consists of diphosphatidylglycerol, phosphatidylglycerol, phosphatidylinositol, phosphatidylinositol mannosides, two unknown aminophospholipids, two phosphoglycolipids, two unknown phospholipids and one unknown lipid. The major fatty acids were iso-C15:0, anteiso-C15:0, iso-C16:0, iso-C16:1 G and anteiso-C17:0. The genomic DNA G+C content was 71.6 mol%. The type strain, KLBMP 1483T (=NBRC 109116T=DSM 45766T) was isolated from surface-sterilized stems of the coastal halophyte Dendranthema indicum (Linn.) Des Moul collected from the coastal region of Nantong, Jiangsu Province, East China.
Accession codes
References
Bèrdy, J. Bioactive microbial metabolites. J. Antibiot. 58, 1–26 (2005).
Qin, S., Xing, K., Jiang, J. H., Xu, L. H. & Li, W. J. Biodiversity, bioactive natural products and biotechnological potential of plant-associated endophytic actinobacteria. Appl. Microbiol. Biotechnol. 89, 457–473 (2011).
Igarashi, Y. et al. Maklamicin, an antibacterial polyketide from an endophytic Micromonospora sp. J. Nat. Prod. 74, 670–674 (2011).
Matsumoto, A., Fukuda, A., Inahashi, Y., Ō mura, S. & Takahashi, Y. Actinoallomurus radicium sp. nov., isolated from the roots of two plant species. Int. J. Syst. Evol. Microbiol. 62, 295–298 (2012).
Koyama, R., Matsumoto, A., Inahashi, Y., Ō mura, S. & Takahashi, Y. Isolation of actinomycetes from the root of the plant, Ophiopogon japonicus, and proposal of two new species, Actinoallomurus liliacearum sp. nov. and Actinoallomurus vinaceus sp. nov. J. Antibiot. 65, 335–340 (2012).
Stackebrandt, E., Rainey, F. A. & Ward-Rainey, N. L. Proposal for a new hierarchic classification system, Actinobacteria classis nov. Int. J. Syst. Bacteriol. 47, 479–491 (1997).
Labeda, D. P., Testa, R. T., Lechevalier, M. P. & Lechevalier, H. A. Glycomyces, a new genus of the Actinomycetales. Int. J. Syst. Bacteriol. 35, 417–421 (1985).
Labeda, D. P. & Kroppenstedt, R. M. Emended description of the genus Glycomyces and description of Glycomyces algeriensis sp. nov., Glycomyces arizonensis sp. nov. and Glycomyces lechevalierae sp. nov. Int. J. Syst. Evol. Microbiol. 54, 2343–2346 (2004).
Evtushenko, L. I., Taptykova, S. D., Akimov, V. N., Semyonova, S. A. & Kalakoutskii, L. V. Glycomyces tenuis sp. nov. Int. J. Syst. Bacteriol. 41, 154–157 (1991).
Gu, Q., Zheng, W. & Huang, Y. Glycomyces sambucus sp. nov., an endophytic actinomycete isolated from the stem of Sambucus adnata Wall. Int. J. Syst. Evol. Microbiol. 57, 1995–1998 (2007).
Qin, S. et al. Glycomyces endophyticus sp. nov., an endophytic actinomycete isolated from the root of Carex baccans Nees. Int. J. Syst. Evol. Microbiol. 58, 2525–2528 (2008).
Qin, S. et al. Glycomyces scopariae sp. nov. and Glycomyces mayteni sp. nov., isolated from medicinal plants in China. Int. J. Syst. Evol. Microbiol. 59, 1023–1027 (2009).
Guan, T. W. et al. Glycomyces halotolerans sp. nov., a novel actinomycete isolated from a hypersaline habitat in Xinjiang, China. Antonie van Leeuwenhoek 100, 137–143 (2011).
Qin, S. et al. Isolation, diversity, and antimicrobial activity of rare actinobacteria from medicinal plants of tropical rain forests in Xishuangbanna, China. Appl. Environ. Microbiol. 75, 6176–6186 (2009).
Shirling, E. B. & Gottlieb, D. Methods for characterization of Streptomyces species. Int. J. Syst. Bacteriol. 16, 313–340 (1966).
Waksman, S. A. The Actinomycetes: a Summary of Current Knowledge, Ronald Press: New York, (1967).
Kelly, K. L. Inter-Society Color Council–National Bureau of Standards Color Name Charts Illustrated with Centroid Colors, US Government Printing Office: Washington, DC, (1964).
Kurup, P. V. & Schmitt, J. A. Numerical taxonomy of Nocardia. Can. J. Microbiol. 19, 1035–1048 (1973).
Williams, S. T., Goodfellow, M. & Alderson, G. Genus Streptomyces Waksman and Henrici 1943, 339AL In: Williams S. T., Sharpe M. E., Holt J. G. (eds) Bergey’s manual of Systematic Bacteriology vol 4, Williams & Wilkins: Baltimore pp 2452–2492 (1989).
Gordon, R. E., Barnett, D. A., Handerhan, J. E. & Pang, C. H. N. Nocardia coeliaca, Nocardia autotrophica and the nocardin strain. Int. J. Syst. Bacteriol. 24, 54–63 (1974).
Qin, S. et al. Abundant and diverse endophytic actinobacteria associated with medicinal plant Maytenus austroyunnanensis in Xishuangbanna tropical rainforest revealed by culturedependent and culture-independent methods. Environ. Microbiol. Rep. 4, 522–531 (2012).
Kim, O. S. et al. Introducing EzTaxon-e: a prokaryotic 16S rRNA Gene sequence database with phylotypes that represent uncultured species. Int. J. Syst. Evol. Microbiol. 62, 716–721 (2012).
Saitou, N. & Nei, M. The neighbor-joining method: a new method for reconstructing phylogenetic tree. Mol. Biol. Evol. 4, 406–425 (1987).
Kluge, A. G. & Farris, F. S. Quantitative phyletics and the evolution of anurans. Syst. Zool. 18, 1–32 (1969).
Felsenstein, J. Evolutionary trees from DNA sequences: a maximum likelihood approach. J. Mol. Evol. 17, 368–376 (1981).
Tamura, K. et al. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28, 2731–2739 (2011).
Felsenstein, J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 39, 783–791 (1985).
Staneck, J. L. & Roberts, G. D. Simplified approach to identification of aerobic actinomycetes by thin-layer chromatography. Appl. Microbiol. 28, 226–231 (1974).
Kawamoto, I., Oka, T. & Nara, T. Cell wall composition of Micromonospora olivoasterospora, Micromonospora sagamiensis, and related organisms. J. Bacteriol. 146, 527–534 (1981).
Uchida, K. & Aida, K. Acyl type of bacterial cell wall: its simple identification by a colorimetric method. J. Gen. Appl. Microbiol. 23, 249–260 (1977).
Collins, M. D., Pirouz, T., Goodfellow, M. & Minnikin, D. E. Distribution of menaquinones in actinomycetes and corynebacteria. J. Gen. Microbiol. 100, 221–230 (1977).
Groth, I. et al. Demetria terragena gen. nov., sp. nov., a new genus of actinomycetes isolated from compost soil. Int. J. Syst. Bacteriol. 47, 1129–1133 (1997).
Minnikin, D. E., Collins, M. D. & Goodfellow, M. Fatty acid and polar lipid composition in the classification of Cellulomonas, Oerskovia and related taxa. J. Appl. Bacteriol. 47, 87–95 (1979).
Mesbah, M., Premachandran, U. & Whitman, W. B. Precise measurement of the G+C content of deoxyribonucleic acid by high-performance liquid chromatography. Int. J. Syst. Bacteriol. 39, 159–167 (1989).
Stackebrandt, E. & Goebel, B. M. Taxonomic note: a place for DNA-DNA reassociation and 16S rRNA sequence analysis in the present species definition in bacteriology. Int. J. Syst. Bacteriol. 44, 846–849 (1994).
Acknowledgements
This research was partially supported by National Natural Science Foundation of China (NSFC No.31370062, 31101502, 31000005), the opening project of the State Key Laboratory of Microbial Resources, Institute of Microbiology, Chinese Academy of Sciences (No.SKLMR-20130601), Qinglan Project of Jiangsu Province (2014) and the Project Funded by the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD).
Author information
Authors and Affiliations
Corresponding authors
Additional information
Supplementary Information accompanies the paper on The Journal of Antibiotics website
Supplementary information
Rights and permissions
About this article
Cite this article
Xing, K., Qin, S., Zhang, WD. et al. Glycomyces phytohabitans sp. nov., a novel endophytic actinomycete isolated from the coastal halophyte in Jiangsu, East China. J Antibiot 67, 559–563 (2014). https://doi.org/10.1038/ja.2014.40
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ja.2014.40