Abstract
A novel angucycline-type antibiotic, warkmycin, was isolated from the culture filtrate of Streptomyces strain Acta 2930. Its chemical structure was elucidated by HR-MS, one-dimensional and 2D NMR experiments. The compound inhibits the growth of Gram-positive bacteria and shows a strong antiproliferative activity against mouse fibroblast cell line NIH-3T3 and human cancer cell lines HepG2 and HT29.
Similar content being viewed by others
Introduction
Our screening of alkaliphilic and alkalitolerant Streptomyces strains isolated from alkaline soil samples collected from the Counties of Durham and Northumberland, UK, resulted in the detection and characterization of novel secondary metabolites with antibiotic and antitumor properties, as exemplified by our reports on lactonamycin Z, pyrocoll, elloxazinones and genoketides.2, 3, 4, 5 HPLC-diode array analysis was applied to the metabolic profiling of extracts from culture filtrates and mycelia of these streptomycetes, following growth in various complex broth media.6 HPLC-diode array evaluation was carried out by means of our in-house HPLC-UV-vis database, which contains nearly 1000 reference compounds, mostly antibiotics.7 Two peaks in the HPLC chromatogram of the culture filtrate extract of Streptomyces strain Acta 2930 grown in oatmeal medium showed characteristic UV-vis spectra. One of them with a retention time of 8.6 min was identified as the antifungal and antitumor antibiotic actiphenol,8 according to an identical retention time and UV-vis spectrum to the reference compound. The second peak had a retention time of 8.3 min and showed an UV-vis spectrum similar, but not identical, to various anthraquinone reference compounds held in the database. This report deals with the classification and fermentation of the producing strain, and with the isolation, structural elucidation and biological activities of warkmycin A (1), a compound that was characterized as a novel antibiotic of the angucycline family. Its structure is shown in Figure 1.
Results
Classification of the producing strain
Strain Acta 2930 produced a pale-yellowish aerial spore mass and a yellow-brown pigmented substrate mycelium on ISP-2 agar;9 diffusible pigments were not detected. Whole-cell hydrolysates of the strain contained LL-diaminopimelic acid while the cellular fatty acid profile was rich in iso- and anteiso-branched chain components with predominant amounts of C15:0 and iso-C16:0. The major isoprenologues were hexa- and octahydrogenated menaquinones with nine isoprene units. All of these properties are consistent with the classification of the strain in the genus Streptomyces,10 an assignment supported by the partial sequencing of the 16S rRNA gene.
Screening, fermentation and isolation
Strain Acta 2930 was one of a collection of filamentous actinomycetes, which were isolated from alkaline soil samples collected from a steel waste tip soil at Consett, County Durham, from a pine forest soil at Hamsterley Forest, County Durham, and from a sand dune at Warkworth, County Northumberland, UK. The strains were grown in various complex media in shake flasks and samples were separated by centrifugation into mycelia and corresponding supernatants. Extracts were analyzed by gradient-mode reversed-phase HPLC and diode array monitoring. Strain Acta 2930 was of interest because of the presence of a characteristic peak in the HPLC chromatogram that was not identified by means of our HPLC-UV-vis database but which showed a similarity in its UV-visible spectrum to those of anthraquinone reference compounds. HPLC-ESI-MS analysis revealed a molecular mass of 1049.5 Da but this did not allow for an unambiguous identification in a DNP database11 search. Consequently, strain Acta 2930 was grown in 10-l stirred tank fermentors using a complex medium. The culture reached a maximal biomass of 8 vol-% after 48 h of incubation, the highest yield of warkmycin (1) was produced in 20 mg l−1 after an incubation period of 64 h. Compound 1 was isolated from the culture filtrate by Amberlite XAD-16 column chromatography followed by ethyl acetate extraction. The raw product was purified by subsequent chromatographic steps on Sephadex LH-20 and Toyopearl HW-40S columns. After concentration to dryness 60 mg of 1 were obtained as a yellow powder. Under acidic separation conditions using 1% CH3COOH in MeOH as eluent, compound 1 was completely decomposed to the deacetylated product 2. Compound 2 was not detected in the fermentation broth.
Structural determination
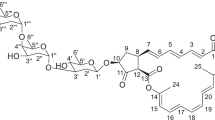
The molecular formula of 1 was established as C50H67NO23 by high-resolution ESI MS. Combination of the 13C-NMR data including DEPT spectrum and a HSQC-NMR spectrum, indicated nine methyl signals, five methylene signals, twenty-one methine signals (including three anomeric carbons) and fifteen quarternary carbons (Table 1). The downfield region of the 1H NMR spectrum of 1 showed a phenolic signal at δH 12.42 and two aromatic protons at δH 7.61 and 7.85; these were complemented by four oxymethine signals and an olefinic signal at δH 5.97. In the 13C-NMR spectrum two quinone carbonyls (δC 186.7 and 190.2) were identified, and the small shift difference of Δδ∼4 p.p.m. was indicative of flanking hydroxy groups in close proximity.12 All these signals were finally assigned to an angucycline core structure. The assignment of the HMBC correlations (Figure 2) confirmed the angucycline scaffold and also proved the attachment of substituents to the angucycline, that is, a methyl group to C3 and two acetoxy groups to C4 and C5, respectively. In addition, three anomeric carbons at δC 99.3, 99.8 and 103.2 were observed in the 13C-NMR spectrum suggesting that compound 1 comprised three glycosidic linkages and therefore the presence of at least three sugar moieties.
A more detailed analysis of one-dimensional (1D) and 2D NMR data of 1 revealed the presence of four sugar moieties, provisionally termed sugars A, B, C and D (Figure 1). Two of the anomeric protons H-1D at δH 4.44, dd, J=1.5, 9.5 Hz (sugar D) and H-1C at δH 4.61, dd, J=1.5, 10.1 Hz (sugar C) showed large coupling constants and thus are indicative of β-glycosidic bound sugars. In contrast, the anomeric proton of sugar A at δH 4.68 revealed a small coupling constant of J=4.3 Hz and hence represents an α-glycosidically linked sugar (Table 1).
The substituents of sugar A were determined by COSY and HMBC correlations as shown in Figure 2. In addition, the comparatively small coupling constant of H-1A at δH 4.68 (J=4.3 Hz), together with three NOE contacts (Figure 3) between the methyl group H-6A and H-3A (both in axial position), between H-4A (ax) and H-5A (eq) and between H-1A and H-6A, revealed axial orientation of the methyl group (H-6A), as well as equatorial orientation of the methoxy group and the carbamoyl group. Comparison of our finding with literature revealed that sugar A is basically a 5-epi-oleandrose. Consequently, sugar A was assigned as 4-O-carbamoyl-α-O-5-epi-oleandrose (4-O-carbamoyl-3-O-methyl-α-O-5-epi-olivose). The position of the substituted carbamoyl group was deduced from HMBC long-range coupling between H-5A and the carbamoyl carbonyl carbon at δC 155.8 and the significant chemical shift observed for the carbon 4A (δC 65.7) if compared with the most similar angucycline, antibiotic P371A113, 14 (4A, δC 73.8). The O-glycosidic linkage of sugar A to C1 of the angucycline system was deduced from the HMBC correlations from H1 to C-1A and from H-1A to C1. Finally, NOESY correlations between H-1A and H1, which reveal an axial position of H1 (Figure 3) at the same time establish the attachment of sugar A to the angucycline core. Furthermore, NOE contacts were observed between H-1 and H-5, which indicates that the two non-aromatic rings are fused in cis-configuration. Together with NOEs for H-5/H-4 and H-5/H-6 this reveals the relative stereochemistry of the angucycline system as shown in Figure 3.
The coupling constant of H-1B (J=10.7 Hz) revealed an axial position of H-1B. Furthermore, NOESY correlations between H-1B and H-3B/H-5B, between H-6B and H-5B showed H-6B and the hydroxy group at H-5B to be equatorial (Figure 3). These findings determined the constitution of sugar B as β-C-olivose bound to C9 of the angucycline system.
In a similar manner, sugar C was established as a β-olivomycose. This was corroborated by NOESY correlations between H-1C and H-5C, between the methyl protons at C3 and H-4C, between H-4C and H-6C (Figure 3). Hence, the coupling constant of H-4C (J=9.8 Hz) reveals axial orientation of H-4C. An HMBC correlation from H-3B to C1 of sugar C established the glycosidic linkage from sugar C to sugar B.
As mentioned before the large coupling constant of H-1D and in combination with the NOE correlation to H-4C are characteristic of an β-glycosidically bound sugar D. NOEs correlations from H-1D to H-3Dax/H-5D and NOEs correlations between the equatorial proton H-3Deq and H-4D, and between H-4D and H-6D revealed equatorial-axial positions of the OH group and the methyl group (H-6D) and therefore determined sugar D as β-amicetose (Figure 3). HMBC correlations from H-1D to C-4C and from H-4C to C-1D revealed an O-glycosidic linkage between sugar D and sugar C. In summary, 1D and 2D NMR data established compound 1 in its relative configuration as novel member of the angucycline antibiotics, named warkmycin.
Compound 2 was obtained during the separation of compound 1 under acidic conditions. The molecular formula of compound 2 was determined as C48H65NO22 by means of high-resolution ESI MS. The mass difference between compound 2 and compound 1 is 42 Da (C2H2O), suggesting the lack of the acetyl group in compound 2. 1H- and 13C-NMR data of 2 showed high similarity to those of compound 1 (Table 1). Nevertheless, slight differences in the spectra were observed. 13C-NMR of compound 2 revealed one signal in the region between 170 and 175 p.p.m. while two signals were observed in the same region of the 13C-NMR spectrum of compound 2. Likewise, the 1H NMR spectrum of compound 2 lacks a singlet signal at δH 2.10, which was assigned to a methyl group in 1. These findings confirmed the absence of the acetyl group in compound 2 attached to C4 of the angucycline core. Besides the 2D NMR data of 2 including COSY, HSQC, HMBC and NOESY experiments were very similar to those of compound 1 and clearly confirmed the suggested structure of 2. In conclusion, the structure of compound 2 termed 4-O-deacetyl-warkmycin was established as the deacetylation product of 1. Table 2 illustrates the physico-chemical data of warkmycin (1) and 4-O-deacetyl-warkmycin (2).
Biological activity
Compounds 1 and 2 showed weak antibacterial activity against the Gram-positive bacterium Staphylococcus lentus; growth inhibition of 23% and 26%, respectively, was observed at a concentration of 100 μM. Both compounds fully inhibited the growth of Bacillus subtilis at the same concentration. Gram-negative bacteria were not susceptible to 1 and 2 at this concentration.
Furthermore, the two compounds exhibited remarkable antiproliferative activity against mouse fibroblast cell line NIH-3T3 and human cancer cell lines HepG2 and HT29. Interestingly; compound 1 exhibited more than a 12-fold potent activity against the fibroblast cell line, whereas both compounds showed similar antiproliferative activity against the cancer cell lines (Table 3).
Discussion
Warkmycin (1) belongs to the family of angucycline antibiotics, which is known to include a vast number of representatives. The angucyclines were reviewed by Rohr and Thiericke15 and defined as compounds with an angular tetracyclic (benz[a]anthracene) moiety and hydrolyzable sugar moieties.15 Angucyclines are distinguished by a variety of biological activities, including antitumor, antibacterial and enzyme inhibitory activities, from which the antitumor activity is the most interesting and most common action. Important representatives of the angucyclines are the landomycins.16 They display a single deoxyoligosaccharide chain of various lengths with up to six carbohydrates, as for landomycin A. Accordingly, the carbohydrate composition of landomycin is a repeated sequence of β-D-olivose-(4→1)-β-D-olivose-(3→1)-α-L-rhodinose attached to a phenolic oxygen of the angucycline core. The biological activities were mainly found to be dependent on the length of the saccharide chain.12 Other examples of angucyclines are the antitumor drugs urdamycin A and B of the aquayamycin-type. Urdamycins display C-glycosylation with three sugar moieties at C9 as a characteristic structural feature.17 Urdamycin A displays two glycosylation positions, at C9 with β-D-olivose-(3→1)-α-L-rhodinose-(4→1)-β-D-olivose and at 12b with L-rhodinose. Because of the presence of an angular oxygen and C- and O-glycosidic moieties, warkmycin (C50H67NO23, 1) also belongs to the aquayamycin-type of angucyclines with angular oxygens and a C-glycosidic moiety.17 Like urdamycin, warkmycin also shows a C-glycosylation at C9 with three sugar moieties. However, the nature and glycosidic linkages of the carbohydrates differ by β-olivose-(4→1)-β-olivomycose-(3→1)-β-amicetose. An even stronger structural relationship to warkmycin is observed for antibiotic P371A1 (C48H66N2O20) from Streptomyces sp. P371,13, 14 but warkmycin differs in the substituents of the sugar units. While the sugar attachment sites to the aglycone are the same, warkmycin has a carbamoyl group substituted at sugar A (α-5-epi-oleandrose) and antibiotic P731A1 has a ureido group at sugar C (D-β-amicetose). Another important difference between warkmycin and antibiotic P371A1 is C-4 of the angucycline moiety. Antibiotic P371A1 contains a methylene group while warkmycin displays a methine group bearing an OAc substituent. Owing to the interesting antiproliferative activity of warkmycin in particular and of angucyclines in general it will be of great interest to further characterize the selectivity of compound 1 against the mouse fibroblast cell line NIH-3T3 by testing other fibroblast cell lines and primary cell cultures.
Methods
Producing strain
A soil sample was collected from an embryo sand dune at Warkworth, Northumberland, UK. Strain Acta 2930 was isolated from the sandy soil sample on starch–casein agar18 adjusted to pH 10.5 (1 M NaOH) and was preserved as a mixture of spores and mycelial fragments in glycerol (20% vol vol−1) at −20 °C. The strain is deposited in the strain collection of the School of Biology at Newcastle University, Newcastle upon Tyne, UK. It was examined for chemotaxonomic and morphological properties known to be of value in streptomycete systematics,19 and by partial 16S rRNA gene sequence analysis.
HPLC screening
The HPLC system consisted of an HP 1090M liquid chromatograph equipped with a diode array detector and an HP Kayak XM 600 ChemStation (Agilent Technologies, Waldbronn, Germany). Sample preparation and chromatographic conditions were performed as described earlier.20 HPLC-ESI-MS analysis was done with an Agilent 1200 HPLC series equipped with a diode array detector and a LC/MSD Ultra Trap System XCT 6330 (Agilent Technologies). Samples (2.5 μl) were injected onto an HPLC column (Nucleosil-100 C-18, 3 μm, 100 × 2 mm; Maisch, Ammerbuch, Germany) and separated by 0.1% aqueous HCOOH as solvent A and 0.06% HCOOH in CH3CN as solvent B by a linear gradient from 10 to 100% B over 15 min at a flow rate of 400 μl min−1.
Fermentation and isolation
Batch fermentations of strain Acta 2930 were performed in 10-l stirred tank fermenters (Biostat E, B. Braun, Melsungen, Germany) in a complex medium that consisted of (per litre tap water) oatmeal 20 g (Neuform, Zarrentin, Germany), and 5 ml trace element solution composed of (per litre deionized water) CaCl2 2H2O 3 g, iron(III) citrate 1 g, MnSO4 H2O 200 mg, ZnCl2 100 mg, CuSO4 5H2O 25 mg, Na2B4O7 10H2O 20 mg, CoCl2 6H2O 4 mg and Na2MoO4 2H2O 10 mg; the pH was adjusted to 7.3 (5 M HCl) before sterilization. The fermenter was inoculated with 5% by volume of a shake flask culture grown in a seed medium at 27 °C in 500 ml-Erlenmeyer flasks with a single baffle for 72 h on a rotary shaker at 120 r.p.m. The seed medium consisted of (per litre tap water) glucose 10 g, glycerol 10 g, oatmeal 5 g (Neuform), soybean meal 10 g (Schoenenberger, Magstadt, Germany), yeast extract 5 g (Ohly Kat, Deutsche Hefewerke, Hamburg, Germany), Bacto casamino acids 5 g (Becton Dickinson, Franklin Lakes, NJ, USA) and CaCO3 1 g. The fermentations were carried out for 64 h with an aeration rate of 0.5 volume air per volume per minute and agitation at 250 r.p.m.
Hyphlo Super-cel (2%; Johns-Manville, Denver, CO, USA) was added to the fermentation broths of two 10-l fermentations, which were separated by multiple sheet filtration into culture filtrate and mycelium. Nineteen liters of culture filtrates were applied to an Amberlite XAD-16 (Rohm and Haas, Frankfurt, Germany) column (resin volume 2 l), and the resin washed with H2O and H2O-MeOH (1:1). Compound 1 was eluted with 100% MeOH and concentrated in vacuo to an aqueous residue (500 ml). The concentrate was adjusted to pH 4.0 and extracted twice with EtOAc (each 100 ml), and the organic extracts combined and concentrated in vacuo to dryness. The crude product (370 mg) was dissolved in MeOH and purified by Sephadex LH-20 (Amersham, Freiburg, Germany) and Toyopearl TSK HW-40 (Tosoh, Stuttgart, Germany) chromatography (each column 90 × 2.5 cm) with MeOH as eluent at a flow rate of 30 ml h−1. After concentration in vacuo to dryness, compound 1 was obtained as a yellow powder with a yield of 60 mg.
Structural determination
High-resolution ESI-MS mass spectra were recorded using a LTQ-Orbitrap XL (Thermo Scientific, Bremen, Germany) coupled to an Agilent 1260 HPLC system (Agilent Technologies). NMR spectra were recorded on a DRX 500 spectrometer (Bruker, Karlsruhe, Germany) equipped with a broadband inverse detection probe head with z gradients at 500 and 125 MHz for 1H and 13C, respectively. CDCl3 was used as a solvent for NMR experiments and chemical shifts were referenced to the solvent peaks (7.24 p.p.m. (1H) and 77.23 p.p.m. (13C)).
Antimicrobial assays
Antimicrobial assays were performed using the Gram-positive bacteria Bacillus subtilis DSM 347 and Staphylococcus lentus DSM 6672, and Gram-negative bacteria Escherichia coli K12 (W1130) and Pseudomonas fluorescens DSM 50090. The growth inhibition was characterized by determining the cell viability using a fluorescence based resazurin assay as described by Schulz et al.21
Cytotoxicity assays
The sensitivity of the cell lines NIH-3T3 (kindly provided by G Rimbach, Universität Kiel, Germany), HepG2 and HT29 to warkmycin was evaluated by monitoring the metabolic activity using the CellTiter-BlueCell Viability Assay (Promega, Mannheim, Germany). Cultivation and the bioassays were performed as described by Schulz et al.21
References
Kalyon, B. et al. Langkocyclines: novel angucycline antibiotics from Streptomyces sp. Acta 3034. J. Antibiot. 66, 609–616 (2013).
Dieter, A. et al. Pyrocoll, an antibiotic, antiparasitic and antitumor compound produced by a novel alkaliphilic Streptomyces strain. J. Antibiot. 56, 639–646 (2003).
Höltzel, A. et al. Lactonamycin Z, an antibiotic and antitumor compound produced by Streptomyces sanglieri strain AK 623. J. Antibiot. 56, 1058–1061 (2003).
Graf, E. et al. Elloxazinones A and B, new aminophenoxazinones from Streptomyces griseus Acta 2871. J. Antibiot. 60, 277–284 (2007).
Fiedler, H.-P. et al. Genoketides A1 and A2, new octaketides and biosynthetic intermediates of chrysophanol produced by Streptomyces sp. AK 671. J. Antibiot. 61, 464–473 (2008).
Goodfellow, M. & Fiedler, H.-P. A guide to successful bioprospecting: informed by actinobacterial systematics. Antonie van Leeuwenhoek 98, 119–142 (2010).
Fiedler, H.-P. Biosynthetic capacities of actinomycetes. 1. Screening for novel secondary metabolites by HPLC and UV-visible absorbance libraries. Nat. Prod. Lett. 2, 119–128 (1993).
Highet, R. J. & Prelog, V. 164. Stoffwechselprodukte von Actinomyceten. 18. Mitteilung, Actiphenol. Helv. Chim. Acta 42, 1523–1526 (1959).
Shirling, E. B. & Gottlieb, D. Methods for characterization of Streptomyces species. Int. J. Syst. Bacteriol. 16, 313–340 (1966).
Kämpfer, P. Genus 1. Streptomyces Waksman and Henrici 1943, 339AL emend Witt and Stackebrandt 1990, 370 emend Wellington, Stackebrandt, Sanders, Wolstrup and Jorgensen 1992, 159 in Bergey’s Manual of Systematic Bacteriology 2nd edn eds Goodfellow M., et al vol. 5 The Actinobacteria Springer: New York, NY, USA, (2012).
Dictionary of Natural Products on DVD Version 20:2 ( CRC Press: London, (2011).
Shaaban, K. A., Stamatkin, C., Damodaran, C. & Rohr, J. 11-Deoxylandomycinone and landomycins X-Z, new cytotoxic angucyclin(on)es from a Streptomyces cyanogenus K62 mutant strain. J. Antibiot. 64, 141–150 (2011).
Uesato, S., Tokunaga, T. & Takeuchi, K. Novel angucycline compound with both antigastrin- and gastric mucosal protective activities. Bioorg. Med. Chem. Lett. 8, 1969–1972 (1998).
Uesato, S. et al. Absolute stereochemistry of gastric antisecretory compound P371A1 and its congener P371A2 from Streptomyces species P371. J. Nat. Prod. 63, 787–792 (2000).
Rohr, J. & Thiericke, R. Angucycline group antibiotics. Nat. Prod. Rep. 9, 103–137 (1992).
Henkel, T., Rohr, J., Beale, J. M. & Schwenen, L. Landomycins, new angucycline antibiotics from Streptomyces sp. I. Structural studies on landomycins A-D. J. Antibiot. 43, 492–503 (1990).
Drautz, H., Zähner, H., Rohr, J. & Zeeck, A. Metabolic products of microorganisms. 234. Urdamycins, new angucycline antibiotics from Streptomyces fradiae. I. Isolation, characterization and biological properties. J. Antibiot. 39, 1657–1669 (1986).
Küster, E. & Williams, S. T. Selective media for the isolation of streptomycetes. Nature 202, 928–929 (1964).
Manfio, G. P., Zakrezewska-Czerwinska, J., Atalan, E. & Goodfellow, M. Towards minimal standards for the description of Streptomyces species. Biotekhnologiya 8, 228–237 (1995).
Nachtigall, J. et al. Atacamycins A-C, 22-membered antitumor macrolactones produced by Streptomyces sp. C38. J. Antibiot. 64, 775–780 (2011).
Schulz, D. et al. Abenquines A-D: aminoquinone derivatives produced by Streptomyces sp. strain DB634. J. Antibiot. 64, 763–768 (2011).
Acknowledgements
Financial support from the European Commission (project ACTAPHARM, 5th framework, QLK3-CT-2001-01783) is gratefully acknowledged. SE Helaly thanks the Egyptian Ministry of Higher Education and Aswan University for a scholarship and support for working at the Technische Universität Berlin. We thank Mr A Kulik, Universität Tübingen, for assistance with the fermentation and HPLC-ESI-MS studies, Ms E Mösker, Technische Universität Berlin, for measurement of the IR data, and Dr T Paululat, Universität Siegen, Germany, for helpful discussions on the manuscript.
Author information
Authors and Affiliations
Corresponding author
Additional information
Art. No. 66 in ‘Biosynthetic Capacities of Actinomycetes’. Art. No. 65: see Manderscheid et al.
Rights and permissions
About this article
Cite this article
Helaly, S., Goodfellow, M., Zinecker, H. et al. Warkmycin, a novel angucycline antibiotic produced by Streptomyces sp. Acta 2930. J Antibiot 66, 669–674 (2013). https://doi.org/10.1038/ja.2013.74
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ja.2013.74
Keywords
This article is cited by
-
Unveiling nature's treasures: actinobacteria from Meghalaya's mining sites as sources of bioactive compounds
Archives of Microbiology (2024)
-
Elmenols C-H, new angucycline derivatives isolated from a culture of Streptomyces sp. IFM 11490
The Journal of Antibiotics (2017)
-
New approaches to antibiotic discovery
Biotechnology Letters (2017)
-
Streptomyces alkalithermotolerans sp. nov., a novel alkaliphilic and thermotolerant actinomycete isolated from a soda lake
Antonie van Leeuwenhoek (2015)
-
New bhimamycins from Streptomyces sp. AK 671
The Journal of Antibiotics (2013)