Abstract
Two new 6,6-spiroacetal polyketides, spirotoamides A (1) and B (2), were isolated from a microbial metabolite fraction library of Streptomyces griseochromogenes JC82-1223 by screening of structurally unique compounds based on a search of spectral database. The fraction library was constructed using a systematic separation method to efficiently discover new metabolites from microbial sources such as actinomycetes and fungi. The structures of 1 and 2 were elucidated by 2D-NMR and mass spectrometric measurements. They belong to a class of polyketides, and contain a 6,6-spiroacetal core structure and a carboxamide group. The biosynthetic pathway of 1 and 2 is discussed in the text.
Similar content being viewed by others
Introduction
Microorganisms, such as actinomycetes and fungi, have a tremendous capacity to produce structurally diverse metabolites1 with disparate activities, rendering them a significant source of pharmaceutical leads and therapeutic agents.2, 3 They are also used as potential bioprobes for chemical biology studies.4, 5 Many secondary metabolites have been reported, but not all have been isolated, nor have their wide-ranging physicochemical properties and low abundance been examined with regard to their potentially useful activities. Thus, we constructed a microbial metabolite fraction library by systematic separation with a spectral database, based on photodiode-array detector attached LC/MS analysis to discover structurally unique metabolites efficiently and rapidly. We discovered and isolated verticilactam6 from S. spiroveticillatus JC-8444, a tautomycin producer,7, 8, 9 and the new fraquinocins I and J10 and 6-dimethylalylindole-3-carbaldehyde11 from S. reveromyceticus SN-593, a reveromycin producer12 by our methodology constructing the fraction library. Also, Bugni et al.13 reported a marine natural products library by HPLC-MS fractionation for rapid drug discovery. These reports have demonstrated the advantage of fraction libraries in isolating and investigating the activities of natural products.
Our microbial fraction library was developed on basic chromatographic techniques, comprizing two-step separation by middle-pressure liquid chromatography (MPLC) and C18-HPLC, yielding 300–400 fractions from a single microbial strain with reproducibility. In addition to many known compounds, the fractions, which contain unidentified minor components, might contain valuable compounds with unique structures or activities or key metabolites from a specific biosynthetic pathway. To take advantage of a fraction library and distinguish new metabolites from known compounds rapidly, a spectral database was constructed, based on photodiode-array-LC/MS analysis, determining the UV absorption and mass spectra of each metabolite within each fraction in the library.
On screening for structurally unique secondary metabolites using the spectral database of the fraction library, we identified two unknown peaks with identical UV and mass spectra in fractions of S. griseochromogenes JC82-1223. The related fractions were purified by a C18-HPLC to yield compounds 1 (4.0 mg) and 2 (1.4 mg), respectively (Table 1). We report the structures of these compounds, designated spirotoamides A (1) and B (2).
Results and Discussion
The molecular formula of compound 1 was determined to be C26H45NO6 by HRESIMS (found: m/z 490.3136 [M+Na]+, calculated for C26H45NO6Na 490.3145). The IR spectrum implied the presence of hydroxyl groups (3332 cm−1) and an amide group (1664 and 1592 cm−1). The 1H NMR spectrum in acetonitrile-d3 suggested that 1 contained five methyl groups and an ethyl group (Supplementary Figure S1). It also contained five exchangeable protons, which were not observed in methanol-d4 (Supplementary Figure S8). Two of the exchangeable protons were observed as broad and symmetrical signals (Table 2), which indicated the presence of a primary amino group; the remaining three exchangeable protons were relatively sharp, suggesting that 1 had three hydroxyl groups. The 13C NMR spectrum showed 26 signals attributable to six methyls, six methylenes, ten methines including four oxygenated and three olefinic carbons, and four quaternary carbons including a carbonyl, an olefinic, an oxygenated sp3, and a distinct acetal carbon at 101.9 p.p.m. (Supplementary Figure S2). These assignments were verified by the 13C DEPT experiment (Supplementary Figure S3) and HSQC spectral data (Table 2; Supplementary Figure S5).
The planar structure of 1 was determined by 2D NMR analysis, as shown in Figure 1. The DQF-COSY spectrum constructed proton spin systems from H-2 to H-3, from H-5 to H-7, from 10-Me to 14-Me with a hydroxyl group at C-13, from H-16 to 19-Me with a hydroxyl and a methyl groups at C-17 and C-18 and from H-1′ to H-2′ (Supplementary Figure S4). The overall structure was constructed using long-range correlations in the HMBC spectrum (Supplementary Figure S6). The HMBC correlations from H-2 and H-3 to C-1 permitted the assignment of the carbonyl carbon as C-1 and the amide group was established by the 13C chemical shift and molecular formula. The ethyl branch at C-4 and the connectivities from C-3 to C-5 were confirmed by the HMBC correlations from H-2′ to C-4, from H-1′ to C-3 and C-5, from H-2 to C-4 and from H-5 to C-3. The HMBC correlations from a methyl group at 1.12 p.p.m. to C-7 and C-8 and from H-7 to C-8 established connectivity between C-7 and C-8 and the methyl branch at C-10. The connectivities from C-8 to C-10 were confirmed by the HMBC correlations from H-9 to C-8, 8-Me, C-10, and C-11 and from 10-Me to C-9. 14-Me, H-14 and H-16 showed HMBC correlations to C-15, establishing the connectivities from C-14 to C-16 through the acetal carbon C-15. A 6,6-spiro-ring system was constructed in consideration of the 13C chemical shift value of C-15 at 101.9 p.p.m.14, 15, 16, 17, 18, 19 and the hydrogen deficiency index of 5, which was also confirmed by the correlations between H-12 and 14-Me, between H-17 and H-19 and between 14-Me and H-16eq in the ROESY spectrum (Supplementary Figure S7). These ROESY correlations suggested that the conformation around C-15 was restricted and they formed spiro-fused tetrahydropyrans. The geometry of the double bonds at Δ2 was assigned as an E-configuration, based on the J of 15.6 Hz in the 1H NMR spectrum. The double bond at Δ4 was also determined as an E-configuration, based on the typical high-field chemical shift value of the C-1′ of 20.5 p.p.m. and the ROESY correlations between H-2 and H-1′, between H-2 and H-2′ and between H-3 and H-5, as shown in Figure 1.
The relative stereochemistry of the 6,6-spiroacetal core structure was analyzed on the basis of the J in the 1H NMR spectrum and ROESY data. To eliminate the influences of the hydroxyl groups on the coupling patterns, NMR spectra were measured in methanol-d4 instead of acetonitrile-d3, and the 1H and 13C NMR chemical shifts were assigned. The key J are summarized in Figure 2a. H-11 was coupled to both protons at C-12, with J of 12.1 and 2.4 Hz, respectively. H-12 at 1.26 p.p.m. was also coupled to H-13 with a J of 12.1 Hz. These data suggested that the tetrahydropyran had a chair conformation and these protons were in axial orientations. 14-Me was assigned to have an axial orientation based on a J of 4.9 Hz between H-13 and H-14, which indicated that H-14 was in an equatorial orientation. Another tetrahydropyran ring was also assigned to have a chair conformation, based on J of 10.8 and 9.7 Hz between H-17 and H-18 and between H-18 and H-19, which suggested that H-17, H-18 and H-19 were in axial orientations. H-16 at 1.17 p.p.m. was assigned to have an axial orientation, based on a J of 10.8 Hz with H-17. These configurations were confirmed by the ROESY data obtained in acetonitrile-d3, as shown in Figure 2b. ROESY correlations between H-11 and H-13 and between H-12 at 1.26 p.p.m. and 14-Me were observed, confirming that the tetrahydropyran had a chair conformation. It indicated that those protons and 14-Me were in axial orientations, respectively, assigning the hydroxyl group at C-13 as an equatorial orientation. The other tetrahydropyran was also confirmed to have a chair conformation, based on the ROESY correlation between H-17 and H-19, which allowed to assign them as axial orientations, respectively. H-17 correlated with H-16 at 1.98 p.p.m. and 18-Me, which also correlated with H-19; these results confirmed that H-16 at 1.98 p.p.m. and H-18 were in equatorial orientations. Based on these results, the structure of 1 was determined as shown in Figure 3.
The compound 2 had the same molecular formula with that of 1 suggesting 2 was a structural isomer of 1. The 1H NMR spectrum was very similar to that of 1, comprizing six methyl signals, including an ethyl group, and five exchangeable protons, two of the exchangeable protons were observed as broad and symmetrical signals, indicating the presence of a primary amino group (Supplementary Figure S9). The remaining three exchangeable protons were however observed as sharp doublets, which suggested that the hydroxyl groups were branched at methine carbons, respectively. Moreover, the 1H NMR spectrum showed the additional signal at 4.47 p.p.m. as a multiplet. The 13C NMR spectrum was nearly identical with that of 1, except for slight differences in chemical shifts of carbon signals around 70 p.p.m. (Supplementary Figure S10). The 13C DEPT experiments (Supplementary Figure S11) and HSQC spectrum (Supplementary Figure S13) revealed that 2 had two additional methine signals, one of which was oxygenated, and lost a methylene signal and a quaternary signal compared with 1 (Table 1). These observations suggested that 2 contained a hydroxyl group at a methine carbon instead of a quaternary carbon like 1. The planar structure including the geometry of double bonds and relative stereochemistry on the tetrahydropyrans was determined by the same manner with those of 1. The newly observed oxygenated methine carbon was assigned to C-6 by the analysis of DQF-COSY data (Supplementary Figure S12). Another new methine carbon was assigned to C-8 with a methyl branch by the DQF-COSY and HMBC data (Supplementary Figure S14). The relative stereochemistry on the tetrahydropyrans was confirmed by the ROESY data (Supplementary Figure S15). Therefore, 2 was a structural isomer of 1 at a substituted position of the hydroxyl group at C-6 as shown in Figure 3.
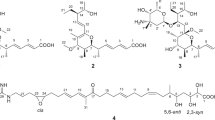
The carbon skeleton of spirotoamides A(1) and B(2) is likely assembled by a type I polyketide synthase, which might catalyze the loading of the acetate unit and the condensation of four malonyl-CoAs, four methylmalonyl-CoAs and one ethylmalonyl-CoA. Based on the knowledge of tautomycin20 and avermectin21 biosyntheses, the presence of a dihydroxyl ketone precursor was expected in the formation of the spiroacetal structure (Figure 4). We recently analyzed the biosynthesis of reveromycin A and identified RevJ responsible for the stereospecific spiroacetal formation from the dihydroxyl ketone precursor.22 To test whether a RevJ homolog existed in the genome of S. griseochromogenes JC82-1223, we performed Southern hybridization analysis using a RevJ probe—the hybridization signal was not found in the spirotoamide-producing strain. Therefore, we speculated that putative dihydroxyl ketone precursor for spirotoamide biosynthesis was non-enzymatically cyclized to form a thermodynamically and electrostatically preferred spiroacetal. It is speculated that the transfer of an amino group to the C-1 carboxyl group by carboxamide synthase and the regiospecific hydroxylation at C-6 or C-8 by P450 enzyme might be involved in the formation of compounds 1 and 2.
Compounds 1 and 2 were subjected to several bioassays in vitro. Their cytotoxic activities against HL-60, HeLa, tsFT210 and tsNRK were evaluated. Also, the antibacterial activities against Escherichia coli HO-141 as a gram-negative bacteria and Staphylococcus aureus 209 as a gram-positive bacteria, and antifungal activities against Candida albicans JCM1542, Aspergillus fumigatus Af293 and Magnaporthe oryzae kita-1 were tested. However, neither compound had any effect at concentrations up to 100 μM.
The spirotoamide-producing strain S. griseochromogenes JC82-1223 synthesizes tautomycetin, which is also a polyketide with a unique 2,3-dialkylmaleic anhydride moiety.23, 24 However, spirotoamides are structurally unrelated to tautomycetin and contain a characteristic 6,6-spiroacetal core structure and carboxamide group, suggesting that our fraction library with spectral database is useful to discover novel microbial metabolites.
Experimental procedure
General experimental procedures
Analytical-grade solvent and reagents were purchased from commercial sources. UV and optical rotations were recorded on a JASCO V-630 BIO spectrophotometer (JASCO International, Tokyo, Japan) and a HORIBA SEPA-300 high-sensitive polarimeter (HORIBA, Kyoto, Japan), respectively. IR spectra were recorded on a HORIBA FT-720 with a DuraSampl IR II ATR instrument (HORIBA). NMR data were obtained at 500 MHz for 1H NMR and 125 MHz for 13C NMR on a JEOL ECP-500 or ECA-500 spectrometer (JEOL, Tokyo, Japan). Chemical shifts (in p.p.m.) were referenced against the residual undeuterated solvent. Mass spectra were obtained on an AB Sciex Qtrap (ESIMS) and JEOL JMS-T100LC (HRESIMS) (AB Sciex, Framingham, MA, USA). LC/MS analysis was performed using a Waters 2996 photodiode-array detector, attached to a 2965 Alliance system, with a Waters Xterra C18-column (2.1 mm i.d. × 150 mm) that was connected to an AB Sciex Qtrap with an ESI probe (Waters, Milford, MA, USA). Teredyne ISCO CombiFlash Companion was used for MPLC (Teredyne ISCO, Lincoln, NE, USA). Preparative HPLC was performed using a Waters 600E pump system with SenshuPak Pegasil ODS column (20 mm i.d. × 250 mm or 10 mm i.d. × 250 mm) (Senshu Scientific, Tokyo, Japan).
Culture condition
S. griseochromogenes JC82-1223 was cultured in a 500 ml of cylindrical flask (K1 flask) containing 70 ml of culture medium (glucose 2%, soluble starch 1%, meat extract 0.3%, dried yeast 2.5%, corn steep liquor 0.3%, K2HPO4 0.005%, NaCl 0.05%, CaCO3 0.05% and MgSO4.7H2O 0.05%) for 3 days at 28 °C on a rotary shaker at 150 r.p.m. 140 ml of each pre-culture was used to inoculate two of 14 l jar-fermentors that contained 7 l of the same culture medium, which were cultured with stirring speed at 100 r.p.m. and an aeration rate of 2 l min−1 for 4 days.
Construction of fraction library
A total 14 l of culture broth was used to construct a microbial metabolites fraction library. The same volume of acetone was added to the whole culture broth and filtered to remove mycelia. The filtrate was evaporated in vacuo to remove acetone, and the remaining aqueous layer was extracted twice with the same volume of ethyl acetate. The organic layer was concentrated to yield 4.2 g of crude extract. The crude extract was subjected to MPLC on 80 g of silica gel packed column with a chloroform/methanol gradient system (methanol 0–100%), yielding seven fractions by the guide of UV chromatogram at 254 nm. Each MPLC fraction was separated by C18-HPLC with a methanol/0.05% aqueous formic acid gradient system into 48 fractions in time constant slices. Each fraction was analyzed by photodiode-array-LC/MS with an acetonitrile/0.05% aqueous formic acid gradient system to collect the UV absorption and mass spectral data for each metabolite. The spectral data were stored in a spectral database.
Isolation
Fraction 26 of the sixth MPLC fraction was purified by C18-HPLC with acetonitrile/water (25:75) to yield compound 1 (4.0 mg). Fraction 27 was subjected to C18-HPLC with acetonitrile/water (24:76) to obtain compound 2 (1.4 mg). Physicochemical properties of 1 and 2 were summarized in Table 1. 1H and 13C NMR chemical shifts of 1 and 2 in acetonitrile-d3 were summarized in Table 2. 1H and 13C NMR chemical shifts of 1 in methanol-d4 were following; δH (multi, J in Hz): 0.88 (Me-14, d, 6.8, 3H), 0.95 (Me-18, d, 6.4, 3H), 1.01 (Me-10, d, 6.8, 3H), 1.05 (H-2′, t, 7.3, 3H), 1.11 (H-18, m), 1.14 (Me-19, d, 6.4, 3H), 1.19 (Me-8, s, 3H), 1.21 (H-16ax, m), 1.24 (H-9, m), 1.34 (H-12ax., br ddd, 12.1, 12.1, 12.1), 1.55 (H-7, br dd, 8.3, 2H), 1.59 (eq., ddd, 12.1, 4.9, 2.4), 1.73 (H-10, qd, 6.8, 2.4), 1.80 (H-9, dd, 14.2, 2.4), 1.83 (H-14, qd, 6.8, 4.9), 2.07 (H-16 eq., dd, 12.7, 4.9), 2.29 (H-6, m, 2H), 2.33 (H-1′, br q, 7.3, 2H), 3.27 (H-19, qd, 9.7, 6.4), 3.29 (H-11, m), 3.52 (H-17, ddd, 10.8, 10.8, 4.9), 4.26 (H-13, ddd, 12.1, 4.9, 4.9), 5.82 (H-5, t, 7.3), 6.00 (H-2, d, 15.6) and 7.08 (H-3, d, 15.6); δC: 7.4 (Me-14), 13.5 (Me-18), 14.0 (C-2′), 18.7 (Me-10), 19.5 (Me-19), 20.7 (C-1′), 24.3 (C-6), 26.8 (Me-8), 31.6 (C-12), 35.1 (C-10), 43.3 (C-16), 43.5 (C-7), 43.6 (C-14), 45.4 (C-9), 46.1 (C-18), 67.1 (C-13), 70.9 (C-17), 71.2 (C-19), 73.5 (C-8), 74.1 (C-11), 102.3 (C-15), 118.3 (C-2), 140.3 (C-4), 141.7 (C-5), 146.4 (C-3) and 171.8 (C-1).
DNA manipulation
DNA isolation and manipulation were performed as described.25 DNA fragments were isolated from agarose using a gel extraction kit (Qiagen, Hilden, Germany). Plasmids were purified using Wizard plus SV Minipreps (Promega, Madison, WI, USA). PCR amplification was performed on a DNAEngine Peltier thermal cycler (BIO-RAD, Hercules, CA, USA).
Isolation of genomic DNA
S. reveromyceticus SN-593 and S. griseochromogenes JC82-1223 were cultured in a K1 flask containing 70 ml of SY medium (0.1% yeast extract (Difco, Becton, Dickinson and Company, Sparks, MD, USA), 0.1% NZ-amine (Wako, Osaka, Tokyo, Japan), and 1% soluble starch; pH 7.0) for 2 days at 28 °C on a rotary shaker at 150 r.p.m. The culture (35 ml) was harvested by centrifugation at 5000 g for 10 min (Allegra X-15R, Beckman coulter, Fullerton, CA, USA). Genomic DNA was isolated as described in Practical Streptomyces Genetics.26
Southern hybridization
Genomic DNA was digested with BglII and applied onto a 0.9% agarose gel and stained with EtBr. The gel was transferred to Hybond-N+ membrane (GE Healthcare, Buckinghamshire, UK). DNA probes for Southern analysis were amplified from template plasmid (pET28b(+)-revJ)22 by KOD-FX DNA polymerase (TOYOBO, Osaka, Japan) at 94 °C for 1 min and 25 cycles of 98 °C for 10 s, 62 °C for 30 s and 68 °C for 1 min. The primers for revJ amplification were RevJ-F: 5′-GTGACCGAGACCGAACAGCTCGAC-3′ and RevJ-R: 5′-TCAGACCCGGGTGAGGTCGACGAC-3′. The amplified DNA was purified by gel extraction (Qiagen) and labeled with AlkPhos Direct Labeling Reagents (GE Healthcare).
Cytotoxicity test
The human promyelocytic leukemia cell line HL-60, human cervical cancer cell line HeLa, mouse breast cancer cell line temperature sensitive (ts) FT210, temperature sensitive normal rat kidney (tsNRK) fibroblasts (RIKEN Cell Bank, Ibaraki, Japan) were cultured in RPMI 1640 (Invitrogen, Carlsbad, CA, USA), supplemented with 10% fetal calf serum, 50 U ml−1 penicillin and 50 μg ml−1 streptomycin, at 37 °C in a humidified atmosphere containing 5% CO2, respectively. Each cell line was seeded in a 96-well culture plate (2 × 104 cells per well for HL-60, 2 × 103 cells per well for HeLa, 1.6 × 104 cells per well for tsFT210 and 1 × 104 cells per well for tsNRK), and then exposed to test compounds for 48 h. Cell growth was measured using Cell Count Reagent SF (Nacalai Tesque, Kyoto, Japan) according to the manufacturer’s instruction. Briefly, 1/100 volume of the WST-8 solution was added to each well and the plates were incubated at 37 °C for 1 h. Then, cell growth was measured as the absorbance at 450 nm on a microplate reader (Perkin Elmer, Waltham, MA, USA).
Antibacterial test
100 μl of aliquot of each culture broth of E. coli HO-141, S. aureus 209, C. albicans JCM1542, A. fumigatus Af293 and M. oryzae kita-1 was transferred to each well in a 96-well culture plate, respectively. Test compounds were added to the culture medium and the plates were incubated at 37 °C for 6 h (E. coli), 24 h (C. albicans), 48 h (A. fumigatus) and 72 h (M. oryzae). Then, the growth of the bacteria or fungus was measured by absorbance at 600 nm. The growth of M. oryzae was estimated by visual inspection.
References
Osada, H. An overview on the diversity of actinomycete metabolites. Actinomycetol. 15, 11–14 (2001).
Newman, D. J. & Cragg, G. M. Natural products as sources of new drugs over the last 25 years. J. Nat. Prod. 70, 461–477 (2007).
Dobson, C. M. Chemical space and biology. Nature 432, 824–828 (2004).
Osada, H. Trends in bioprobe research. in Bioprobes (ed. Osada, H.) 1–14 (Springer, Berlin, 2000).
Osada, H. Chemical biology based on small molecule-protein interaction. Protein targeting with small molecules. in Chemical Biology Techniques and Applications (ed. Osada, H.) 1–10 (Wiley, New Jersey, 2009).
Nogawa, T. et al. Verticilactam, a new macrolactam isolated from a microbial metabolite fraction library. Org. Lett. 12, 4564–4567 (2010).
Cheng, X. et al. A new antibiotic, tautomycin. J. Antibiot. 40, 907–909 (1987).
Magae, J., Watanabe, C., Osada, H., Cheng, X. C. & Isono, K. Induction of morphological change of human myeloid leukemia and activation of protein kinase C by a novel antibiotic, tautomycin. J. Antibiot. 41, 932–937 (1988).
Cheng, X. C., Ubukata, M. & Isono, K. The structure of tautomycin, a dialkylmaleic anhydride antibiotic. J. Antibiot. 43, 809–819 (1990).
Panthee, S. et al. Furaquinocins I and J: novel polyketide isoprenoid hybrid compounds from Streptomyces reveromyceticus SN-593. J. Antibiot. 64, 509–513 (2011).
Takahashi, S. et al. Biochemical characterization of a novel indole prenyltransferase from Streptomyces sp. SN-593. J. Bacteriol. 192, 2839–2851 (2010).
Osada, H., Koshino, H., Isono, K., Takahashi, H. & Kawanishi, G. Reveromycin A, a new antibiotic which inhibits the mitogenic activity of epidermal growth factor. J. Antibiot. 44, 259–261 (1991).
Bugni, T. M. et al. Marine natural product libraries for high-throughput screening and rapid drug discovery. J. Nat. Prod. 71, 1095–1098 (2008).
Seto, H. & Otake, N. The 13C-NMR spectra of polyether antibiotics and some empirical rules for structural studies of polyether antibiotics. Heterocycles 17, 555–580 (1982).
Koshino, H., Takahashi, H., Osada, H. & Isono, K. Reveromycins, new inhibitors of eukaryotic cell growth. J. Antibiot. 45, 1420–1427 (1992).
Höltzel, A., Kempter, C., Metzger, J. W. & Jung, G. Spirofungin, a new antifungal antibiotic from Streptomyces violaceusniger Tü 4113. J. Antibiot. 51, 699–707 (1998).
Igarashi, Y., Iida, T., Yoshida, R. & Furumai, T. Pteridic acids A and B, novel plant growth promoters with auxin-like activity from Streptomyces hygroscopicus TP-A0451. J. Antibiot. 55, 764–767 (2002).
Niggemann, J. et al. Spirangien A and B, highly cytotoxic and antifungal spiroketals from the myxobacterium Sorangium cellulosum: isolation, structure elucidation and chemical modifications. Eur. J. Org. Chem. 24, 5013–5018 (2005).
Li, J. et al. Virgatolides A–C, benzannulated spiroketals from the plant endophytic fungus Pestalotiopsis virgatula. Org. Lett. 13, 2670–2673 (2011).
Li, W., Ju, J., Rajski, S. R., Osada, H. & Shen, B. Characterization of the tautomycin biosynthetic gene cluster from Streptomyces spiroverticillatus unveiling new insights into dialkylmaleic anhydride and polyketide biosynthesis. J. Biol. Chem. 283, 28607–28617 (2008).
Ikeda, H., Nonomiya, T. & Omura, S. Organization of biosynthetic gene cluster for avermectin in Streptomyces avermitilis: analysis of enzymatic domains in four polyketide synthases. J. Ind. Microbiol. Biotechnol. 27, 170–176 (2001).
Takahashi, S. et al. Reveromycin A biosynthesis uses RevG and RevJ for stereospecific spiroacetal formation. Nat. Chem. Biol. 7, 461–468 (2011).
Cheng, X. C. et al. A new antibiotic, tautomycetin. J. Antibiot. 42, 141–144 (1989).
Cheng, X. C., Ubukata, M. & Isono, K. The structure of tautomycetin, a dialkylmaleic anhydride antibiotic. J. Antibiot. 43, 890–896 (1990).
Sambrook, J. & Russell, D. W. Molecular Cloning: A Laboratory Manual 3rd edn (Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y., 2001).
Kieser, T., Bibb, M. J., Buttner, M. J., Chater, K. F. & Hopwood, D. A. Practical Streptomyces Genetics (John Innes Foundation, Norwich, UK, 2000).
Acknowledgements
We thank Drs T Nakamura and Y Hongo (RIKEN) for HRESIMS measurements. We also thank Drs H Koshino, H Hirota and T Shimizu (RIKEN) for useful discussions on the structures, Mr H Konno (RIKEN) for construction of spectral database, Dr Y Futamura for biological activity tests and Dr M Ueki (RIKEN) for helpful assistance. This work was supported in part by the Target Protein Research Program from the Ministry of Education, Culture, Sports, Science and Technology of Japan.
Author information
Authors and Affiliations
Corresponding author
Additional information
Supplementary Information accompanies the paper on The Journal of Antibiotics website
Supplementary information
Rights and permissions
About this article
Cite this article
Nogawa, T., Takahashi, S., Okano, A. et al. Spirotoamides A and B, novel 6,6-spiroacetal polyketides isolated from a microbial metabolite fraction library. J Antibiot 65, 123–128 (2012). https://doi.org/10.1038/ja.2011.121
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ja.2011.121
Keywords
This article is cited by
-
Structure and biological activity of Metarhizin C, a stereoisomer of BR-050 from Tolypocladium album RK17-F0007
The Journal of Antibiotics (2019)
-
Kinanthraquinone, a new anthraquinone carboxamide isolated from Streptomyces reveromyceticus SN-593-44
The Journal of Antibiotics (2018)
-
Wakodecalines A and B, new decaline metabolites isolated from a fungus Pyrenochaetopsis sp. RK10-F058
The Journal of Antibiotics (2018)
-
Competition and co-regulation of spirotoamide and tautomycetin biosynthesis in Streptomyces griseochromogenes, and isolation and structural elucidation of spirotoamide C and D
The Journal of Antibiotics (2017)
-
Opantimycin A, a new metabolite isolated from Streptomyces sp. RK88-1355
The Journal of Antibiotics (2017)