Abstract
Rapamycin and its analogs are clinically important macrolide compounds produced by Streptomyces hygroscopicus. They exhibit antifungal, immunosuppressive, antitumor, neuroprotective and antiaging activities. The core macrolactone ring of rapamycin is biosynthesized by hybrid type I modular polyketide synthase (PKS)/nonribosomal peptide synthetase systems primed with 4,5-dihydrocyclohex-1-ene-carboxylic acid. The linear polyketide chain is condensed with pipecolate by peptide synthetase, followed by cyclization to form the macrolide ring and modified by a series of post-PKS tailoring steps. The aim of this review was to outline past and recent advances in the biosynthesis and regulation of rapamycin, with an emphasis on the distinguished contributions of Professor Demain to the study of rapamycin. In addition, this article describes the biological activities as well as mechanism of action of rapamycin and its derivatives. Recent attempts to improve the productivity of rapamycin and generate diverse rapamycin analogs through mutasynthesis and mutagenesis are also introduced, along with some future perspectives.
Similar content being viewed by others
Introduction
Rapamycin (1; Figure 1), produced by Streptomyces hygroscopicus, is a 31-membered macrocyclic natural product exhibiting various biological and pharmacological activities, including antifungal,1 immunosuppressive,2 antitumor,3 neuroprotective4 and antiaging activities.5 Like the structurally related FK506, another Streptomyces metabolite with potent immunosuppressive activities,6 rapamycin has attracted interest for the clinical treatment of organ transplant rejection2, 7 and autoimmune disease.8 Even though rapamycin and FK506 have a similar polyketide backbone and share a common cellular receptor, the FK506-binding protein (FKBP), these immunosuppressants act through different mechanisms in cells. FK506 inhibits interleukin 2-mediated T-cell proliferation by blocking the Ca2+/calcineurin-dependent transcriptional activation of the genes responsible for growth, whereas rapamycin prevents growth-promoting cytokine signaling by interacting with a mammalian target of rapamycin (mTOR) instead of calcineurin (Figure 2).9
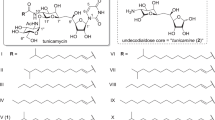
Structure of rapamycin and its analogs. (1) Rapamycin, (2) prolylrapamycin, (3) 32-desmethylrapamycin (WAY-125286), (4) 32-desmethoxyrapamycin (WAY-24688), (5) deferolimus (AP23573), (6) everolimus (RAD001), (7) temsirolimus (CCI779), (8) WYE-592, (9) ILS-920, (10) 4-hydroxyprolylrapamycin, (11) 4-hydroxyprolyl-26-demethoxyrapamycin, (12) 4-thiarapamycin and (13) 9-deoxo-5-sulfoxyrapamycin. (14), (15) and (16) rapamycin analogs generated by feeding pseudostarter units such as cyclohexanecarboxylic acid (CHC), cyclohex-1-ene-carboxylic acid and cycloheptanecarboxylic acid, respectively.
Mechanism of action of rapamycin. Inhibition of mTOR signaling by rapamycin regulates cell proliferation, angiogenesis and cell metabolism. mTOR, mammalian target of rapamycin; IRS1, insulin receptor substrate-1; PI3K, phosphoinositide 3-kinase; PDK1, phosphoinositide-dependent protein kinase 1; AKT (PKB), protein kinase B; gβL, GTPase β-subunit-like protein; raptor, regulatory-associated protein of TOR; rictor, rapamycin-insensitive companion of TOR; deptor, disheveled, Egl-10, pleckstrin (DEP) domain containing mTOR interacting protein; protor, proline-rich protein; 4E-BPs, eukaryotic translation initiation factor 4E-binding proteins; PRAS40, proline-rich AKT substrate of 40 kDa; p70S6K, ribosomal p70 receptor; S6K, S6 kinase; ASK1, apoptosis signal-regulating kinase 1; JNK, c-JUN N-terminal kinase.
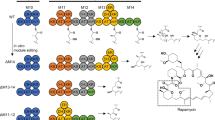
In the early 1990s, a study on the biosynthesis of immunosuppressant rapamycin was begun by Demain's research group at Massachusetts Institute of Technology.10 Classical feeding experiments with isotope-labeled precursors provided evidence that rapamycin is formed through a polyketide pathway and its macrolactone ring is derived from acetate, propionate and methionine.11 The biosynthesis of the polyketide chain is initiated by the incorporation of a shikimate-derived 4,5-dihydroxycyclohex-1-ene carboxylic acid (DHCHC) starter unit, which in turn is elongated by 14 condensation steps with 7 acetate units and 7 propionate units (Figure 3).11 The chain growth is then completed by the attachment of a pipecolate moiety to the linear polyketide product through a nonribosomal peptide synthetase, which also catalyzes the macrolactone ring closure to generate pre-rapamycin (Figure 3).12, 13, 14 In addition, formation of rapamycin requires further post-polyketide synthase (PKS) modification steps, including oxidations catalyzed by cytochrome P450 monooxygenases, and O-methylations by S-adenosylmethionine-dependent methyltransferases.12, 13, 15
Since the discovery of rapamycin PKS,12 Demain and his colleagues have studied the effect of nutrients on rapamycin production in S. hygroscopicus, and reported that good rapamycin production can be achieved when adding a low concentration of ammonium in the absence of phenylalanine and methionine, which are normally used as the nitrogen sources during the fermentation.16, 17 Supplementation with ferrous salt and a limitation of phosphate and magnesium salts are also required to increase rapamycin productivity.18 Furthermore, exogenous shikimic acid was found to double rapamycin production,19 whereas prolylrapamycin (2), usually produced in trace amounts, became the major product when adding proline to the fermentation, due to competition between proline and endogenous pipecolic acid.20 Interestingly, when rapamycin analogs, such as prolylrapamycin, 32-desmethylrapamycin (WAY-125286; 3) and 32-desmethoxyrapamycin (WAY-24688; 4), were examined for their antifungal activities, all were more active than a conventionally used antifungal drug amphotericin B, although they were less active than rapamycin.21 Fang et al.22 also observed an interesting effect of the naturally coproduced elaiophylin and nigericin on the antifungal activity of rapamycin, where both of them enhanced the anti-Candida albicans activity of rapamycin.
On the basis of Demain's pioneering works, more recent efforts to increase rapamycin productivity and generate novel biologically active analogs of rapamycin have focused on precursor-directed biosynthesis, mutagenesis and mutasynthesis. For example, the Biotica group (Cambridge, UK) revealed various positive regulatory elements in rapamycin biosynthesis,23 the Wyeth group (Madison, NJ, USA) modified the pipecolate ring of rapamycin and Leadlay and coworkers24 synthesized several rapamycin analogs with modifications to the cyclohexyl moiety through genetic engineering and mutasynthesis.
Recently, Graziani25 provided a review article on the advances in the field of rapamycin biosynthesis and neuroprotective activity of rapamycin analogs. There have been also a large number of thorough reviews on the potential of using rapamycin for cancer, organ transplantation and autoimmune disorders. This review summarizes the role of rapamycin and its biosynthesis, while also describing previous and current attempts to regulate rapamycin production by nutritional control, mutagenesis for improved production and mutasynthesis for the generation of novel analogs. Precursor engineering and combinatorial biosynthesis strategies are also introduced for further improving the efficiency of rapamycin production and generating diverse useful rapamycin analogs.
Biological activities of rapamycin and its analogs
When rapamycin was first isolated from soil microbes collected on Easter Island, it attracted attention due to its antifungal properties against human pathogenic yeasts, Candida spp.1, 26 Rapamycin provided lower minimum inhibitory concentrations against eight clinical isolates of Candida when compared with other antifungal agents, such as candicidin, nystatin and amphotericin B.1 Rapamycin was also found to be very active in the treatment of systemic candidosis in mice when compared with amphotericin B, and previous reports on the rapamycin–FKBP complex in yeast showed that the formation of the complex is essential for the antifungal activity of rapamycin.27, 28
Although rapamycin was originally studied as an antifungal agent, it was soon identified as a potent immunosuppressant and is currently in use and under development as treatment in transplant medicine.2, 29 Rapamycin was approved in the United States in 1999 (and 2000 in Europe) for the prevention of acute rejection in combination with cyclosporine and steroids. A large number of groups have already shown the ability of rapamycin to diminish pathology in various animal model immune diseases.9 Interestingly, rapamycin inhibits the chronic/acute organ rejection process that is not suppressed by cyclosporine and FK506.30, 31 It has been suggested that these immunosuppressive effects of rapamycin result from its ability to inhibit proliferation by interfering with the function of mTOR.32 When rapamycin–FKBP complex interacts with mTOR, this blocks the cell-cycle progression of T cells in the mid-to-late G1 phase, thereby suppressing the T-cell proliferation induced by cross-linking of the T-cell receptors, antigenic peptides or cytokines such as interleukins.33 The effects of rapamycin are not only limited to T-cell proliferation, but also include the proliferation of B lymphocytes.34
In addition to its antifungal and anti-immunosuppressive effects, rapamycin and its derivatives, such as deferolimus (AP23537; 5), everolimus (RAD001; 6) and temsirolimus (CCl779; 7) shown in Figure 1, have been recently identified as potent anticancer agents.35, 36 It has also been shown that rapamycin and its analogs effectively suppress cellular proliferation and angiogenesis in a number of human malignancies.37, 38 More recently, it was reported that eukaryotic translation initiation factor 4E-binding proteins activation in vivo by rapamycin (Figure 2) prevented dopaminergic neuron loss,39 which is characteristic of the neurodegenerative disorder parkinsonism.40 Along with their neuroprotective activities, rapamycin and its analogs have been also shown to stimulate neurite outgrowth.41 A structure-based drug design approach used by Graziani et al.25 showed that semisynthetic analogs of rapamycin, WYE-592 (8) and ILS-920 (9) (Figure 1), could be promising candidates for an in vivo neuroprotection/neurogeneration study. Furthermore, Harrison et al.5 showed that rapamycin extended the median and maximal life span of both male and female mice beginning at 600 days old in separate studies simultaneously replicated at three different test sites. Feeding rapamycin to male and female mice beginning at 270 days old also increased survival, which may have resulted from the postponement of death from cancer, retarding the mechanism of aging or both.
Figure 2 shows the action mechanism of mTOR, the serine/threonine kinase that controls the cellular processes of growth, proliferation, transcription, protein biosynthesis and ribosomal biogenesis.42 mTOR exists in two distinct protein complexes referred to as mTOR complex 1 and mTOR complex 2. Rapamycin-sensitive mTOR complex 1 consists of mTOR, a GTPase β-subunit-like protein, deptor and regulatory associated protein of TOR (raptor), whereas mTOR complex 2 includes mTOR, deptor, a GTPase β-subunit-like protein, a rapamycin-insensitive companion of TOR (rictor) and protor.43 The inactivation of mTOR complex 1 kinase activity by rapamycin results in the inhibition of the activities of ribosomal S6 kinase and the eukaryotic translation initiation factor 4E-binding proteins, which have roles in ribosome biogenesis and protein translation, respectively. In contrast, apoptosis and autophagy are stimulated by rapamycin.32, 44
Biosynthesis of rapamycin
The initial precursor incorporation experiments using 13C-labeled acetate and propionate showed that the macrolide ring of rapamycin is biosynthesized from seven acetate and seven propionate units. The three O-methyl groups of rapamycin are derived from methionine.11 Later, several competitive incorporation studies using labeled precursors proved that the heterocyclic ring originates from pipecolic acid, which is formed from lysine (Figure 3).45 The feeding of 13C-labeled shikimic acid prepared from 13C-glucose also elucidated that the cyclohexane ring in rapamycin is derived from the shikimic acid pathway.46
The rapamycin biosynthetic genes from S. hygroscopicus have been identified by hybridization with the PKS genes for erythromycin biosynthesis.12 The rapamycin PKS consists of three multifunctional enzymes (RapA, RapB and RapC) with a total of 14 modules: modules 1–4 in RapA (900 kDa) involved in polyketide chain initiation and extension, modules 5–10 in RapB (1070 kDa) required for chain elongation up to C16 and modules 11–14 in RapC (660 kDa) responsible for the termination of the polyketide portion of the macrolactone ring. The organization of the rapamycin gene cluster is shown in Figure 3. The loading module located in the N-terminal region of RapA contains the characteristic domain, ATP-dependent carboxylic acid/CoA ligase and enoyl reductase.15 The CoA ligase domain activates the unusual starter unit DHCHC, forming acyladenylate, and then transfers the activated acyl group directly to the β-ketoacyl-acyl carrier protein (ACP) synthase domain of module 1. The adjoining enoyl reductase domain is proposed to reduce the starter unit after it has linked to RapA.15 The subsequent modules carry a set of domains for β-ketoacyl-ACP synthase, acyltransferase and ACP, which catalyze polyketide chain elongation, including auxiliary β-keto processing domains, ketoreductase, dehydratase and enoyl reductase. Most β-ketoacyl-ACP synthase, acyltransferase and ACP domains in rapamycin PKS are highly conserved, although the detailed sequences of the acyltransferase domains appear to be divided into two groups based on the substrate specificity of the extender unit, (2S)-malonyl-CoA or (2S)-methylmalonyl-CoA.15 Except for module 14, the dehydratase and ketoreductase domains are present in all rapamycin PKS. Meantime, the enoyl reductase, ketoreductase and dehydratase domains in module 3, and dehydratase and ketoreductase domains in module 6 are presumed to be inactive, even though the sequences are indistinguishable from those of other active domains.15
Other genes responsible for rapamycin biosynthesis have been also identified by sequencing beyond the PKS region.12 The chain synthesis is terminated by a specialized protein encoded by rapP, designated a pipecolate-incorporating enzyme, which inserts pipecolate moiety into the polyketide backbone and catalyzes subsequent cyclization.12 The evidence that pipecolate incorporation and macrolactone ring closure are carried out by pipecolate-incorporating enzyme was obtained by a nucleotide sequence analysis and disruption of rapP from S. hygroscopicus.47 A comparison of the deduced amino-acid sequence of rapP with those of authentic peptide synthetases, such as GrsT from gramicidin S,48 TycF from tyrocidine49 and SrfD from surfactin biosynthetic gene cluster,50 revealed the presence of highly conserved motifs for ATP binding, aminoacyl adenylate formation, peptide bond formation and substrate transfer.47 When rapP was disrupted from the chromosome of S. hygroscopicus, rapamycin production was significantly reduced. Furthermore, recombinant pipecolate-incorporating enzyme expressed in the heterologous host Streptomyces coelicolor CH999 was labeled with 14C-pipecolate, indicating that the pipecolate was covalently bound to the enzyme through a thioester linkage and consequently it was phosphopantetheinylated.47 Pipecolate, a direct precursor for the heterocyclic ring of rapamycin, is biosynthesized from lysine by rapL encoding a lysine cyclodeaminase. The role of RapL was shown based on the chromosomal disruption experiment in S. hygroscopicus.20 In addition, heterologous expression and purification of recombinant RapL from Escherichia coli also characterized the in vitro enzymatic activity of RapL.51
Modifications of the 31-memebered lactone ring by oxidation and O-methylation are thought to be the final steps in the biosynthesis of rapamycin. Recently, the Sheridan group identified the functions of several gene products involved in post-PKS tailoring by biotransformation and gene complementation.52 The predicted functions of the post-PKS tailoring enzymes corresponding to the conversion of pre-rapamycin into rapamycin are shown in Figure 3. RapJ and RapN encode cytochrome P450 monooxygenases that catalyze the specific oxidation step in C9 and C27, respectively, of the pre-rapamycin macrolide. RapO shares a sequence homology with known ferredoxins that are required for the activity of cytochrome P450 enzymes. The deduced gene products of rapM, rapQ and rapI are similar to S-adenosylmethionine-dependent O-methyltransferases, which carry out methylation at the hydroxyl groups of C16, C27 and C39, respectively, in the rapamycin molecule.15, 52
Regulation of rapamycin biosynthesis
Despite accumulated information on the rapamycin biosynthetic pathway, most of the nutritional control and regulatory factors remain unknown. Demain's group examined the effect of carbon53 and nitrogen sources54 on the biosynthesis of rapamycin in S. hygroscopicus. Among 35 carbon sources, including polysaccharides, oligosaccharides, monosaccharides and organic acids, 12 carbon sources were found to support the growth of the producer strain and rapamycin production in a chemically defined medium. In particular, seven carbon sources, that is, fructose, mannose, galactose, inositol, mannitol, xylose and cellobiose, were found to stimulate rapamycin production. Although the best combination of carbon sources was 2% fructose and 0.5% mannose, a combination of 1.5 g l−1 aspartic acid, 0.5 g l−1 arginine and 0.5 g l−1 histidine was an effective mixture of nitrogen sources for the growth of S. hygroscopicus and rapamycin production.53
On the basis of these preliminary results, Demain's team examined additional 18 amino acids under fermentation conditions supplemented with aspartic acid, arginine and histidine.54 As a result, rapamycin formation was significantly increased by lysine, yet decreased by phenylalanine and methionine. The lysine stimulation was presumably due to its role as a precursor of pipecolic acid, which is incorporated into the pre-rapamycin, whereas the negative impact of phenylalanine on the formation of shikimic acid, a rapamycin precursor, had an interfering effect on the rapamycin production.16 It would appear that methionine represses the generation of rapamycin, where methionine inhibits at least one O-methyltransferase involved in rapamycin biosynthesis and S-adenosylmethionine synthetase.55 Among the six non-amino acids (ammonium sulfate, ammonium nitrate, ammonium chloride, ammonium citrate, urea and potassium nitrate), the addition of ammonium sulfate stimulated rapamycin formation.54 Demain's research group reported on the effects of phosphate, ammonium, magnesium and iron on rapamycin biosynthesis, and found that rapamycin production was increased when adding FeSO4 up to 0.36 mM, greater than that required for growth, whereas phosphate (K2HPO4), ammonium (NH4Cl) and magnesium salts (MgSO4·7H2O) interfered with rapamycin formation at concentrations optimal for growth.18
The addition of exogenous shikimate resulted in an approximately twofold enhanced rapamycin production.19 As a result, it has been predicted that shikimic acid serves as a precursor for the biosynthesis of the trisubstituted cyclohexane ring (DHCHC) outside the macrolactone from the acetate and propionate units.11 However, the previously observed inhibition of rapamycin biosynthesis by phenylalanine treatment16 was not recovered even by shikimate.19
Demain and his coworkers also evaluated the relationship between rapamycin production and a simulated microgravity environment that has only received limited attention. The growth of S. hygroscopicus under simulated microgravity conditions provided by a rotating-wall bioreactor led to pellet formation during cell growth, a decrease in the dry cell weight and interference in rapamycin production. An attempt to reduce the pellet formation by adding Teflon beads to the rotating-wall bioreactor proved to be unsuccessful as regards increasing the rapamycin production.56, 57
A number of regulatory protein families, as well as physical and chemical factors including nutritional sources and microgravity, can affect a broad range of physiological processes involved in secondary metabolite production. Therefore, targeted genetic engineering of the regulatory system can be an alternative approach to improve the productivity of useful natural products. From a sequence analysis study, several open-reading frames from the rapamycin biosynthetic gene cluster have been identified having a potential regulatory function.13 RapR and RapS, which appear to be translationally coupled proteins, have sequence similarities with bacterial response sensor and regulator proteins.13 Meanwhile, RapH, RapG and RapY have been shown to contain a helix-turn-helix motif for DNA binding.13 Although RapY exhibits sequence similarity to repressors of antibiotic export such as ActII from actinorhodin58 and TcmR from tetracenomycin gene clusters,59 the sequences of RapG are similar to those of the positive regulatory proteins SoxS and Rob from E. coli.13 The N-terminal region of RapH contains a potential ATP-binding motif matching the canonical P-loop consensus.13 A sequence analysis also revealed that both RapH and RapG contain the rare leucine codon TTA, which has been proposed to serve as part of the regulatory mechanism of secondary metabolites in Streptomyces.60
Recently, the Biotica group showed that RapH and RapG have important roles as positive regulators in rapamycin biosynthesis.23 Overexpression of rapH and rapG under the control of the ActII-ORF4/PactI activator/promoter in S. hygroscopicus enhanced the production of rapamycin by approximately 27–55% and 20–32%, respectively. The ActII-ORF4/PactI expression system, which is a robust activator/promoter expression system that has been widely used for many actinomycetes, including S. hygroscopicus, was used to minimize the potential self-regulatory interference of RapH and/or RapG and other endogenous regulatory genes.61 It was also observed that the introduction of an additional copy of rapH and rapG under the control of native promoter region of S. hygroscopicus increased rapamycin formation by 40% on average, when compared to that in the wild-type strain. Deletion experiments of both genes from the chromosome of S. hygroscopicus also confirmed the positive regulatory role of RapG and RapH in rapamycin biosynthesis. Subsequent complementation with rapH and rapG resulted in a restoration of antibiotic production, underlying their essential role.23
Mutagenesis and mutasynthesis
Although rapamycin is a valuable product with various pharmacological activities, the productivity of rapamycin in wild-type S. hygroscopicus is low. Therefore, further improvement of the rapamycin titer and generation of diverse derivatives of rapamycin in S. hygroscopicus has been imperative for its commercial development and clinical applications.62 In an attempt to improve rapamycin productivity, fermentation medium optimization as described in the previous section,16, 18, 53, 54 in addition to classical strain improvement,63 protoplasts-related techniques64 and high-throughput screening method65 have been developed. Demain's research group obtained a higher rapamycin-producing mutant from the protoplasts of S. hygroscopicus FC904 mutants treated with 1 μg ml−1 of gentamicin, widely used aminoglycosides, where an HPLC analysis showed that the rapamycin titer for this mutant (139 mg l−1) was 60% higher than that for the parent strain. An enhanced productivity of rapamycin (194 mg l−1; 124% higher than that of starting strain) was also obtained when exposing the spores of the S. hygroscopicus FC904 strain to gentamicin and N-methyl-N′-nitro-N-nitrosoguanidine.63
Another attempt to generate high-yield rapamycin-producing strains was based on the protoplast-related techniques such as protoplasts mutation, intra- and interspecies fusion. When using mutation by UV, N-methyl-N′-nitro-N-nitrosoguanidine, diethyl sulfates treatment or fusion of different parental protoplasts of S. hygroscopicus, progress was not achieved. However, the combination of interspecies protoplasts fusion and one round of genome shuffling generated a high-yield rapamycin producer with an outstanding yield of 445 mg l−1.64 Furthermore, as a result of the protoplast-related mutation and fusion experiments, it was observed that nearly all the high-yield rapamycin-producing strains shared a similar phenotype: round colonies with wrinkled edges and abundant yellow aerial mycelia.
Most traditional screening techniques performed in Erlenmeyer flasks require a large amount of material and cultivation process, thereby limiting the wide application of strain improvement strategies.66 Yet, a high-throughput screening method for isolating high-yield rapamycin-producing strains was recently developed by Xu et al.,65 where more than 7000 colonies were screened and 10 high-yielding strains were isolated based on 96-well microtiter-plates cultivation and an ‘agar plug’ method. As a result, one mutant among the 10 isolates yielded 420 mg l−1 rapamycin, which was double the productivity of the parent strain used in the submerged fermentation process.65
Mutasynthesis through genetic engineering is another useful approach for generating novel derivatives of rapamycin. In a previous study, the Leadlay research group at Cambridge University reported that when wild-type S. hygroscopicus was grown in a rich medium supplemented with proline, it produced not only rapamycin but also prolylrapamycin (2; Figure 1), a rapamycin analog in which pipecolate is replaced by proline in the macrolactone ring. This indicates that RapP, which is responsible for incorporating the pipecolate precursor into the macrocycle, has a relaxed substrate specificity.20 They also found that novel rapamycins containing 4-hydroxyprolyl moieties were biosynthesized in an engineered strain of S. hygroscopicus mutant bearing a deletion of rapL, where the deduced product is able to convert lysine to pipecolic acid. Also, when fermentations of rapL-deleted S. hygroscopicus mutants constructed by Ö31-phage-mediated gene replacement were added with 4-hydroxyproline, 4-hydroxyprolylrapamycin (10) and 4-hydroxyprolyl-26-demethoxyrapamycin (11) were produced (Figure 1).20
Meantime, another group showed that the use of nipecotic acid inhibited the biosynthesis of pipecolate, thereby enhancing the incorporation of other pipecolate analogs into the rapamycin molecule. Thus, 4-thiarapamycin (12; Figure 1) and 9-deoxo-5-sulfoxyrapmycin (13; Figure 1) were obtained from cultures of S. hygroscopicus supplemented with sulfur-containing pipecolate analogs, (S)-1,4-thiazane-3-carboxylic acid and (S)-1,3-thiazane-4-carboxylic acid, respectively.67 However, in an FKBP-binding assay, both of these analogs displayed a significantly weaker binding affinity than rapamycin, supporting the previous report that the pipecolate-containing region of the rapamycin molecule is critical for the binding of rapamycin to FKBP.68, 69
The addition of exogenous unnatural starter units also provided another way of generating structurally modified rapamycin analogs. When carboxylic acids such as cyclohexanecarboxylic acid, cyclohex-1-ene-carboxylic acid and cycloheptanecarboxylic acid in place of the natural starter unit DHCHC were fed to cultures of an engineered strain of S. hygroscopicus MG2-10 in which rapKIJMNOQL is deleted from the rapamycin biosynthetic gene cluster, the resulting mutants gave pre-rapamycin analogs.70 This study was based on the precursor-directed biosynthesis carried out by Lowden et al.24 in which the feeding of alternative starter units, such as cyclohexanecarboxylic acid, cyclohex-1-ene-carboxylic acid and cycloheptanecarboxylic acid, to the fermentation of rapamycin-producing S. hygroscopicus led to the production of new rapamycin derivatives (compounds 14, 15 and 16; Figure 1).
Perspectives
Although conventional strain improvement has enhanced the production level of rapamycin without genetic information on target gene clusters, it is still laborious and time consuming. However, metabolic engineering of the precursor pathways required for rapamycin biosynthesis may be able to control the carbon flux and lead to improved production of rapamycin and its derivatives. The extender units for rapamycin biosynthesis are malonyl-CoA and methylmalonyl-CoA, which are commonly used for polyketide biosynthesis. Malonyl-CoA is usually derived from the carboxylation of acetyl-CoA by an acetyl-CoA carboxylase.71 Meanwhile, it has been suggested that methylmalonyl-CoA can be produced through several pathways, including the isomerization of succinyl-CoA, carboxylation of propionyl-CoA, catabolism of valine, CoA ligation of methylmalonate and conversion of acetyl-CoA through crotonyl-CoA-dependent pathway, as shown in Figure 4.72 Recently, it was reported that the production of FK506 in Streptomyces clavuligerus CKD119 was improved by enhancing the supply of the biosynthetic precursor methylmalonyl-CoA.73 Thus, if precursor pathway engineering is coupled with the traditional mutagenesis described herein, an efficient platform for rapamycin production could be achieved.
Proposed pathways for increasing supply of rapamycin precursors. Engineering the diverse biosynthetic pathway of malonyl-CoA and methylmalonyl-CoA enhances the rapamycin productivity. PCC, propionyl-CoA carboxylase; MatB, malonyl/methylmalonyl-CoA ligase; MCM, methylmalonyl-CoA mutase; CCR, crotonyl-CoA carboxylase/reductase; MeaA, putative coenzyme-B12-dependent mutase; ACC, acyl-CoA carboxylase.
Combinatorial biosynthesis using the modular genetic architecture of rapamycin PKS may also offer great potential possibility for engineering enzymes to produce novel drug candidates. It has been reported that two novel tetraketides and four octaketides were biosynthesized by combining the rapamycin PKS module with another macrolide erythromycin PKS.74 Therefore, ongoing studies of the details and mechanisms involving rapamycin biosynthesis and enzymology will be able to generate multiple rapamycin analogs with potential pharmaceutical application.
References
Sehgal, S. N., Baker, H. & Vézina, C. Rapamycin(AY-22 989), a new antifungal antibiotic. II. Fermentation, isolation and characterization. J. Antibiot. 28, 727–733 (1975).
Calne, R. Y. et al. Rapamycin for immunosuppression in organ allografting. Lancet 120, 443–444 (1989).
Douros, J. & Suffness, M. New antitumor substances of natural origin. Cancer Treat. Rev. 8, 63–87 (1981).
Steiner, J. P. et al. Neurotrophic actions of nonimmunosuppressive analogues of immunosuppressive drugs FK506, rapamycin and cyclosporin A. Nat. Med. 3, 421–428 (1997).
Harrison, D. E. et al. Rapamycin fed late in life extends lifespan in genetically heterogeneous mice. Nature 460, 392–395 (2009).
Motamedi, H., Cai, S. J., Shafiee, A. & Elliston, K. O. Structural organization of a multifunctional polyketide synthase involved in the biosynthesis of the macrolide immunosuppressant FK506. Eur. J. Biochem. 244, 74–80 (1997).
Kahan, B. D. Efficacy of sirolimus compared with azathioprine for reduction of acute renal allograft rejection: a randomised multicentre study. The Rapamune US Study Group. Lancet 356, 194–202 (2000).
Baeder, W. L., Sredy, J., Sehgal, S. N., Chang, J. Y. & Adams, L. M. Rapamycin prevents the onset of insulin-dependent diabetes mellitus (IDDM) in NOD mice. Clin. Exp. Immunol. 89, 174–178 (1992).
Dumont, F. J. & Su, Q. Mechanism of action of the immunosuppressant rapamycin. Life Sci. 58, 373–395 (1996).
Demain, A. L. Pickles, pectin, and penicillin. Annu. Rev. Microbiol. 58, 1–42 (2004).
Paiva, N. L., Demain, A. L. & Roberts, M. F. Incorporation of acetate, propionate, and methionine into rapamycin by Streptomyces hygroscopicus. J. Nat. Prod. 54, 167–177 (1991).
Schwecke, T. et al. The biosynthetic gene cluster for the polyketide immunosuppressant rapamycin. Proc. Natl Acad. Sci. USA 92, 7839–7843 (1995).
Molnár, I. et al. Organisation of the biosynthetic gene cluster for rapamycin in Streptomyces hygroscopicus: analysis of genes flanking the polyketide synthase. Gene 169, 1–7 (1996).
Gregory, M. A. et al. Isolation and characterization of pre-rapamycin, the first macrocyclic intermediate in the biosynthesis of the immunosuppressant rapamycin by S. hygroscopicus. Angew. Chem. Int. Ed. Engl. 43, 2551–2553 (2004).
Aparicio, J. F. et al. Organization of the biosynthetic gene cluster for rapamycin in Streptomyces hygroscopicus: analysis of the enzymatic domains in the modular polyketide synthase. Gene 169, 9–16 (1996).
Cheng, Y. R., Fang, A. & Demain, A. L. Effect of amino acids on rapamycin biosynthesis by Streptomyces hygroscopicus. Appl. Microbiol. Biotechnol. 43, 1096–1098 (1995).
Kim, W. S., Xu, L., Souw, D., Fang, A. & Demain, A. L. An unexpected inhibitory effect of rapamycin against germination of spores of Bacillus brevis strain Nagano. J. Antibiot. 55, 650–654 (2002).
Cheng, Y. R., Hauck, L. & Demain, A. L. Phosphate, ammonium, magnesium and iron nutrition of Streptomyces hygroscopicus with respect to rapamycin biosynthesis. J. Ind. Microbiol. 14, 424–427 (1995).
Fang, A. & Demain, A. L. Exogenous shikimic acid stimulates rapamycin biosynthesis in Streptomyces hygroscopicus. Folia. Microbiol. (Praha) 40, 607–610 (1995).
Khaw, L. E., Böhm, G. A., Metcalfe, S., Staunton, J. & Leadlay, P. F. Mutational biosynthesis of novel rapamycins by a strain of Streptomyces hygroscopicus NRRL 5491 disrupted in rapL, encoding a putative lysine cyclodeaminase. J. Bacteriol. 180, 809–814 (1998).
Wong, G. K., Griffith, S., Kojima, I. & Demain, A. L. Antifungal activities of rapamycin and its derivatives, prolyrapamcyin, 32-desmethylrapamycin, and 32-desmethoxyrapamycin. J. Antibiot. 51, 487–491 (1998).
Fang, A., Wong, G. K. & Demain, A. L. Enhancement of the antifungal activity of rapamycin by the coproduced elaiophylin and nigericin. J. Antibiot. 53, 158–162 (2000).
Kuscer, E et al. Roles of rapH and rapG in positive regulation of rapamycin biosynthesis in Streptomyces hygroscopicus. J. Bacteriol. 189, 4756–4763 (2007).
Lowden, P. A., Böhm, G. A., Metcalfe, S., Staunton, J. & Leadlay, P. F. New rapamycin derivatives by precursor-directed biosynthesis. Chembiochem 5, 535–538 (2004).
Graziani, E. I. Recent advances in the chemistry, biosynthesis and pharmacology of rapamycin analogs. Nat. Prod. Rep. 26, 602–609 (2009).
Vézina, C., Kudelski, A. & Sehgal, S. N. Rapamycin (AY-22 989), a new antifungal antibiotic I. Taxonomy of the producing Streptomyces and isolation of the active principle. J. Antibiot. 28, 721–726 (1975).
Heitman, J., Movva, N. R., Hiestand, P. C. & Hall, M. N. FK506-binding protein proline rotamase is a target for the immunosuppressive agent FK506 in Saccharomyces cerevisiae. Proc. Natl Acad. Sci. USA 88, 1948–1952 (1991).
Koltin, Y. et al. Rapamycin sensitivity in Saccharomyces cerevisiae is mediated by a peptidyl-prolyl cis-trans isomerase related to human FK506-binding protein. Mol. Cell. Biol. 11, 1718–1723 (1991).
Watson, C. J. E. Sirolimus (rapamycin) in clinical transplantation. Transplant. Rev. 15, 165–177 (2001).
Meiser, B. M., Billingham, M. E. & Morris, R. E. Effects of cyclosporin, FK506, and rapamycin on graft-vessel disease. Lancet 338, 1297–1298 (1991).
Gregory, C. R., Huie, P., Billingham, M. E. & Morris, R. E. Rapamycin inhibits arterial intimal thickening caused by both alloimmune and mechanical injury. Its effect on cellular, growth factor, and cytokine response in injured vessels. Transplantation 55, 1409–1418 (1993).
Sehgal, S. N. Sirolimus: its discovery, biological properties, and mechanism of action. Transplant. Proc. Rev. 35, 7S–14S (2003).
Terada, N. et al. Rapamycin blocks cell cycle progression of activated T cells prior to events characteristic of the middle to late G1 phase of the cycle. J. Cell. Physiol. 154, 7–15 (1993).
Aagaard-Tillery, K. M. & Jelinek, D. F. Inhibition of human B lymphocyte cell cycle progression and differentiation by rapamycin. Cell. Immunol. 156, 493–507 (1994).
Petroulakis, E., Mamane, Y., Le Bacquer, O., Shahbazian, D. & Sonenberg, N. mTOR signaling: implications for cancer and anticancer therapy. Br. J. Cancer 94, 195–199 (2006).
Faivre, S., Kroemer, G. & Raymond, E. Current development of mTOR inhibitors as anticancer agents. Nat. Rev. Drug Discov. 5, 671–688 (2006).
Kapoor, A. & Figlin, R. A. Targeted inhibition of mammalian target of rapamycin for the treatment of advanced renal cell carcinoma. Cancer 115, 3618–3630 (2009).
Gulhati, P. et al. Targeted inhibition of mammalian target of rapamycin signaling inhibits tumorigenesis of colorectal cancer. Clin. Cancer Res. 15, 7207–7216 (2009).
Tain, L. S. et al. Rapamycin activation of 4E-BP prevents parkinsonian dopaminergic neuron loss. Nat. Neurosci. 12, 1129–1135 (2009).
Farrer, M. J. Genetics of Parkinson disease: paradigm shifts and future prospects. Nat. Rev. Genet. 7, 306–318 (2006).
Parker, E. M., Monopoli, A., Ongini, E., Lozza, G. & Babij, C. M. Rapamycin, but not FK506 and GPI-1046, increases neurite outgrowth in PC12 cells by inhibiting cell cycle progression. Neuropharmacology 39, 1913–1919 (2000).
Dowling, R. J., Topisirovic, I., Fonseca, B. D. & Sonenberg, N. Dissecting the role of mTOR: lessons from mTOR inhibitors. Biochim. Biophys. Acta. 1804, 433–439 (2010).
Sarbassov, D. D. et al. Prolonged rapamycin treatment inhibits mTORC2 assembly and Akt/PKB. Mol. Cell 22, 159–168 (2006).
Abraham, R. T. & Eng, C. H. Mammalian target of rapamycin as a therapeutic target in oncology. Expert. Opin. Ther. Targets 12, 209–222 (2008).
Paiva, N. L., Demain, A. L. & Roberts, M. F. The immediate precursor of the nitrogen-containing ring of rapamycin is free pipecolic acid. Enzyme Microb. Technol. 15, 581–585 (1993).
Pavia, N. L., Roberts, M. F. & Demain, A. L. The cyclohexane moiety of rapamycin is derived from shikimic acid in Streptomyces hygroscopicus. J. Ind. Microbiol. 12, 423–428 (1993).
König, A. et al. The pipecolate-incorporating enzyme for the biosynthesis of the immunosuppressant rapamycin—nucleotide sequence analysis, disruption and heterologous expression of rapP from Streptomyces hygroscopicus. Eur. J. Biochem. 247, 526–534 (1997).
Krätzschmar, J., Krause, M. & Marahiel, M. A. Gramicidin S biosynthesis operon containing the structural genes grsA and grsB has an open reading frame encoding a protein homologous to fatty acid thioesterases. J. Bacteriol. 171, 5422–5429 (1989).
Mootz, H. D. & Marahiel, M. A. The tyrocidine biosynthesis operon of Bacillus brevis: complete nucleotide sequence and biochemical characterization of functional internal adenylation domains. J. Biotechnol. 179, 6843–6850 (1997).
Steller, S. et al. Initiation of surfactin biosynthesis and the role of the SrfD-thioesterase protein. Biochemistry 43, 11331–11343 (2004).
Gatto, Jr G. J., Boyne, M. T. 2nd, Kelleher, N. L. & Walsh, C. T. Biosynthesis of pipecolic acid by RapL, a lysine cyclodeaminase encoded in the rapamycin gene cluster. J. Am. Chem. Soc. 128, 3838–3847 (2006).
Gregory, M. A. et al. Rapamycin biosynthesis: elucidation of gene product function. Org. Biomol. Chem. 4, 3565–3568 (2006).
Kojima, I., Cheng, Y. R., Mohan, V. & Demain, A. L. Carbon source nutrition of rapamycin biosynthesis in Streptomyces hygroscopicus. J. Ind. Microbiol. 14, 436–439 (1995).
Lee, M. S., Kojima, I. & Demain, A. L. Effect on nitrogen source on biosynthesis of rapamycin by Streptomyces hygroscopicus. J. Ind. Microbiol. Biotechnol. 19, 83–86 (1997).
Kim, W. S., Wang, Y., Fang, A. & Demain, A. L. Methionine interference in rapamycin production involves repression of demethylrapamycin methyltransferase and S-adenosylmethionine synthetase. Antimicrob. Agents Chemother. 44, 2908–2910 (2000).
Fang, A., Pierson, D. L., Mishra, S. K. & Demain, A. L. Growth of Steptomyces hygroscopicus in rotating-wall bioreactor under simulated microgravity inhibits rapamycin production. Appl. Microbiol. Biotechnol. 54, 33–36 (2000).
Demain, A. L. & Fang, A. Secondary metabolism in simulated microgravity. Chem. Rec. 1, 333–346 (2001).
Fernández-Moreno, M. A., Caballero, J. L., Hopwood, D. A. & Malpartida, F. The act cluster contains regulatory and antibiotic export genes, direct targets for translational control by the bldA tRNA gene of Streptomyces. Cell 66, 769–780 (1991).
Guilfoile, P. G. & Hutchinson, C. R. Sequence and transcriptional analysis of the Streptomyces glaucescens tcmAR tetracenomycin C resistance and repressor gene loci. J. Bacteriol. 174, 3651–3658 (1992).
Leskiw, B. K., Mah, R., Lawlor, E. J. & Chater, K. F. Accumulation of bldA-specified tRNA is temporally regulated in Streptomyces coelicolor A3(2). J. Bacteriol. 175, 1995–2005 (1993).
Kieser, T., Bibb, M. J., Buttner, M. J., Chater, K. F. & Hopwood, D. A. Practical Streptomyces Genetics (The John Innes Foundation, Norwich, 2000 ).
Adrio, J. L. & Demain, A. L. Genetic improvement of processes yielding microbial products. FEMS Microbiol. Rev. 30, 187–214 (2006).
Cheng, Y. R., Huang, J., Qiang, H., Lin, W. L. & Demain, A. L. Mutagenesis of the rapamycin producer Streptomyces hygroscopicus FC904. J. Antibiot. 54, 967–972 (2001).
Chen, X. et al. Generation of high-yield rapamycin-producing strains through protoplasts-related techniques. Appl. Microbiol. Biotechnol. 83, 507–512 (2009).
Xu, Z. N., Shen, W. H., Chen, X. Y., Lin, J. P. & Cen, P. L. A high-throughput method for screening of rapamycin-producing strains of Streptomyces hygroscopicus by cultivation in 96-well microtiter plates. Biotechnol. Lett. 27, 1135–1140 (2005).
Duetz, W. A. et al. Methods for intense aeration, growth, storage, and replication of bacterial strains in microtiter plates. Appl. Environ. Microbiol. 66, 2641–2646 (2000).
Ritacco, F. V. et al. Production of novel rapamycin analogs by precursor-directed biosynthesis. Appl. Environ. Microbiol. 71, 1971–1976 (2005).
Graziani, E. I. et al. Novel sulfur-containing rapamycin analogs prepared by precursor-directed biosynthesis. Org. Lett. 5, 2385–2388 (2003).
Van Duyne, G. D., Standaert, R. F., Karplus, P. A., Schreiber, S. L. & Clardy, J. Atomic structure of FKBP-FK506, an immunophilin-immunosuppressant complex. Science 252, 839–842 (1991).
Gregory, M. A. et al. Mutasynthesis of rapamycin analogues through the manipulation of a gene governing starter unit biosynthesis. Angew. Chem. Int. Ed. Engl. 44, 4757–4760 (2005).
Bianchi, A. et al. Identification of an isozymic form of acetyl-CoA carboxylase. J. Biol. Chem. 265, 1502–1509 (1990).
Chan, Y. A., Podevels, A. M., Kevany, B. M. & Thomas, M. G. Biosynthesis of polyketide synthase extender units. Nat. Prod. Rep. 1, 90–114 (2009).
Mo, S., Ban, Y. H., Park, J. W., Yoo, Y. J. & Yoon, Y. J. Enhanced FK506 production in Streptomyces clavuligerus CKD1119 by engineering the supply of methylmalonyl-CoA precursor. J. Ind. Microbiol. Biotechnol. 36, 1473–1482 (2009).
Rowe, C. J et al. Engineering a polyketide with a longer chain by insertion of an extra module into the erythromycin-producing polyketide synthase. Chem. Biol. 8, 475–485 (2001).
Acknowledgements
This work was supported by National Research Foundation of Korea Grant funded by the Korean Government (20090083305, 20090083153 and 20100001487), Seoul R&BD Program (KU080657M0209721) and Technology Development Program for Fisheries, Ministry for Food, Agriculture, Forestry and Fisheries, Republic of Korea.
Author information
Authors and Affiliations
Corresponding author
Additional information
This paper is contributed to the special issue of The Journal of Antibiotics celebrating Dr Arnold Demain's 60 years of scientific career.
Rights and permissions
About this article
Cite this article
Park, S., Yoo, Y., Ban, YH. et al. Biosynthesis of rapamycin and its regulation: past achievements and recent progress. J Antibiot 63, 434–441 (2010). https://doi.org/10.1038/ja.2010.71
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ja.2010.71
Keywords
This article is cited by
-
Rapamycin golden jubilee and still the miraculous drug: a potent immunosuppressant, antitumor, rejuvenative agent, and potential contributor in COVID-19 treatment
Bioresources and Bioprocessing (2022)
-
Crossregulation of rapamycin and elaiophylin biosynthesis by RapH in Streptomyces rapamycinicus
Applied Microbiology and Biotechnology (2022)
-
Genome-based analysis for the bioactive potential of Streptomyces yeochonensis CN732, an acidophilic filamentous soil actinobacterium
BMC Genomics (2020)
-
Genetic manipulation of secondary metabolite biosynthesis for improved production in Streptomyces and other actinomycetes
Journal of Industrial Microbiology and Biotechnology (2016)
-
Characterization of negative regulatory genes for the biosynthesis of rapamycin in Streptomyces rapamycinicus and its application for improved production
Journal of Industrial Microbiology & Biotechnology (2015)