Abstract
Glutamine-dependent amidotransferase (Gn-AT) catalyzes the transfer of the amido nitrogen of glutamine to an acceptor substrate to produce glutamate and an aminated product. Although most Gn-ATs are involved in biosyntheses of primary metabolites, some are important in the synthesis of secondary metabolites. Recently, a Gn-AT (NspN) was discovered in the biosynthetic pathway of 4-hydroxy-3-nitorosobenzamide in Streptomyces murayamaensis. NspN converts 3-amino-4-hydroxybenzoic acid (3,4-AHBA) to 3-amino-4-hydroxybenzamide. Here, we report the amino-acceptor substrate specificity of NspN. NspN could use several benzoic acid derivatives as amino-acceptor substrates to produce corresponding benzamides and catalyze the amidation of several carboxylate-type phenylpropanoids, such as p-coumaric acid, cinnamic acid and caffeic acid. NspN showed the highest activity toward its natural substrate 3,4-AHBA among the substrates examined. NspN and related bacterial Gn-ATs may be useful in developing combinatorial biosynthetic strategies for benzamide derivatives, which could, in turn, be used as therapeutic agents for a variety of diseases.
Similar content being viewed by others
Introduction
The amido group on the side chain of glutamine is often used as a nitrogen source for biological molecules by glutamine-dependent amidotransferases (Gn-ATs). Gn-ATs catalyze the transfer of the amido nitrogen of glutamine to an acceptor substrate to produce glutamate and an aminated product. This reaction is composed of two half reactions catalyzed by two independent active sites within the enzyme.1, 2 The first reaction is the hydrolysis of glutamine and the transfer of ammonia to the other active site. The second reaction is the activation of an acceptor substrate and transfer of the ammonia to form the aminated product. Each reaction proceeds in two independent domains of the enzyme, a glutaminase domain for nitrogen transfer at the N terminus and an ATP-dependent synthetase (or synthase) domain for amination at the C terminus. Ammonia travels from the glutaminase domain to the synthetase domain through an ammonia channel.2 Gn-ATs are classified primarily into two subfamilies, classes I and II, according to their glutaminase domains.3 Glutaminase domains of class I Gn-ATs consist of ∼200 amino-acid residues and have a cysteine protease-type catalytic triad, formed by Cys, His, and Gln. In contrast, class II Gn-ATs have a glutaminase domain in which the N-terminal Cys is the only catalytic residue. Although the glutaminase domain uses glutamine as a common nitrogen donor, the synthetase domain exhibits amination activity with a variety of acceptors to produce several metabolites, most of which are primary metabolites, such as amino acids, amino sugars, purine and pyrimidine nucleotides, and coenzymes. There are only a few examples of Gn-ATs that are involved in secondary metabolic processes. PhzH catalyzes the amidation of phenazine-1-carboxylic acid to phenazine-1-carboxamide in the biosynthetic pathway of phenazine-1-carboxamide in Pseudomonas aeruginosa PAO14 and Pseudomonas chlororaphis PCL1391.5 OxyD and SsfD are involved in the biosynthesis of oxytetracycline in Streptomyces rimosus6 and tetracycline SF2575 in Streptomyces sp. SF2575,7 respectively. PcsA and PcsB are polyene carboxamide synthases in the polyene amide biosynthesis pathway in Streptomyces diastaticus8 and the AB-400 biosynthetic pathway in Streptomyces RGU5.3,9 respectively. TsrC is involved in the biosynthesis of thiostrepton in Streptomyces laurentii.10
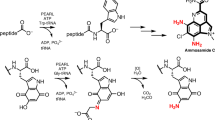
Recently, we determined the entire biosynthetic pathway of 4-hydroxy-3-nitrosobenzamide (4,3-HNBAm) in Streptomyces murayamaensis in which NspN, a class II Gn-AT, plays an essential role. NspN converts 3-amino-4-hydroxybenzoic acid (3,4-AHBA), synthesized from L-aspartic-4-semialdehyde and dihydroxyacetone phosphate by NspI and NspH, into 3-amino-4-hydroxybenzamide, which is then oxidized by NspF, a novel nitroso-forming o-aminophenol oxidase, to yield 4,3-HNBAm (Figure 1).11 NspN amidates 3,4-AHBA, but not 4-hydroxy-3-nitorosobenzoic acid (4,3-HNBA), indicating that amidation precedes oxidation in the conversion of 3,4-AHBA to 4,3-HNBAm.11
Biosynthetic pathway of 4-hydroxy-3-nitrosobenzamide (4,3-HNBAm). 3-Amino-4-hydroxybenzoic acid (3,4-AHBA) is formed from L-aspartic-4-semialdehyde and dihydroxyacetone phosphate by NspI and NspH. NspN catalyzes the conversion of 3,4-AHBA to 3-amino-4-hydroxybenzamide (3,4-AHBAm) through amidation of the carboxyl group of 3,4-AHBA. Oxidation of the amino group of 3,4-AHBAm is then catalyzed by NspF to yield 4,3-HNBAm.
The current study further examined the amino-acceptor substrate specificity of NspN. Several benzamide derivatives are used as therapeutic agents for psychiatric diseases (for example, sulpiride, amisulpride, sultopride12 and nemonapride13), as analgesic (for example, salicylamide14) and as gastrointestinal agents (for example, metoclopramide15 and itopride16). Knowledge of the amino-acceptor substrate specificity of NspN would significantly aid the development of combinatorial biosyntheses using benzamide synthetases.
Materials and methods
Chemicals
4-Hydroxy-3-nitrosobenzoic acid (>94% purity) was enzymatically synthesized using recombinant NspF as described previously.11 L-Aspartic-4-semialdehyde was synthesized according to the methods described by Black and Wright.17 ATP was purchased from Sigma (St Louis, MO, USA). L-Glutamine and L-asparagine were purchased from Nippon Rika, Tokyo, Japan. MgCl2 was purchased from Kokusan Chemical, Tokyo, Japan. Other compounds were purchased from Wako Chemicals, Osaka, Japan.
Enzyme assays of NspN-H activity
The His6-tag fusion NspN protein (NspN-H) was expressed and purified as described previously.11 A standard reaction mixture (50 μl) consisted of 5 mM aromatic substrate, 5 mM ATP, 10 mM L-glutamine, 10 mM MgCl2, 100 mM HEPES [4-(2-hydroxyethyl) piperazine-1-ethanesulfonic acid sodium salt]-NaOH buffer (pH 7.5) and NspN-H (3.3–4.9 μg). The mixture was preincubated without enzyme for 10 min at 30 °C, and the reaction was started by addition of the enzyme. After incubation for 60 min at 30 °C, the reaction was stopped by the addition of 50 μl of 5% (v/v) trifluoroacetic acid. The reaction products were identified by LC-ESIMS as described below.
A Shodex ODP2 HP-4E column (4.6 × 250 mm, Showa Denko K.K., Tokyo, Japan) was used for both HPLC and LC-ESIMS to measure the products of 3,4-AHBA and 4-amino-3-hydroxybenzoic acid catalysis. LC-ESIMS was performed in positive/negative mode on an HPLC system (model 1100 series, Agilent Technologies, Santa Clara, CA, USA) equipped with a mass spectrometer (Bruker HCT plus, Bruker Daltonics, Bremen, Germany). The substrates and products were eluted with a linear gradient of 20–100% (v/v) acetonitrile containing 0.1% (v/v) acetic acid for 35 min at a flow rate of 0.3 ml min−1.
The reaction products of the other benzoic acid derivatives and carboxylate-type phenylpropanoids were analyzed on a Senshu Pak DOCOSIL-B columns (2.0 × 200 mm for LC-ESIMS, 3.0 × 250 mm for HPLC; Senshu Scientific Co., Tokyo, Japan). LC-ESIMS was performed in positive/negative mode as described above. Both substrates and products were eluted with a linear gradient of 0–100% (v/v) acetonitrile containing 0.1% (v/v) acetic acid for 25 min at a flow rate of 0.3 ml min−1.
For enzymatic property analysis, production of 3-amino-4-hydroxybenzamide (3,4-AHBAm) from 3,4-AHBA by the NspN reaction was measured by HPLC. For HPLC, both 3,4-AHBA and 3,4-AHBAm were eluted with a linear gradient of 20–100% (v/v) acetonitrile containing 0.1% (v/v) acetic acid for 20 min at a flow rate of 0.5 ml min−1. Eluted compounds were detected by monitoring the elute absorbance at 270 nm with a L-2450 diode array detector (Hitachi, Tokyo, Japan).
For substrate specificity analysis, the enzymatic activity toward each substrate was measured by a spectrophotometric method. The standard 50-μl reaction mixture was incubated for 150 min at 30 °C, and the reaction was stopped by the addition of 10 μl of 5 M HCl. After incubation for 30 min at room temperature, the mixture was neutralized by the addition of 10 μl of 5 M NaOH and 130 μl of 230 mM HEPES-NaOH (pH 7.5), and mixed with pyrophosphate reagent (Sigma). The presence of pyrophosphate, which was produced from ATP by the NspN reaction, was detected by measuring the solution absorbance at 340 nm. The reaction mixture containing boiled NspN-H was also examined as a negative control.
Only a fraction of the total available substrate (less than 10%) was converted by NspN-H. Thus, the conversion rate was regarded as the apparent activity of NspN-H toward each substrate. Each assay was run in triplicate, and the results reported here are mean values.
Results and discussion
Enzymatic properties of NspN
NspN is composed of two well-conserved protein motifs commonly associated with asparagine synthetase: a class II glutamine amidotransferase signature in the N-terminal glutaminase domain and an asparagine synthetase signature in the C-terminal synthetase domain. These features indicate that NspN should use glutamine as an amino donor and that ATP and Mg2+ should be required for the activation of an amino acceptor. To examine the amino-acceptor substrate specificity of NspN, a recombinant NspN protein11 (NspN-H) having a structure of NspN-His6 was produced in Streptomyces lividans and purified to homogeneity via metal affinity chromatography and anion exchange chromatography.11 As expected, NspN-H showed no activity in the absence of glutamine, ATP or Mg2+, and asparagine could not be used as an amino donor (data not shown). NspN-H was stable at pH 7.5–10 (at 16 °C for 18 h) and below 30 °C (at pH 7.5 for 1 h; Figure 2). These ranges are typical for a cytoplasmic enzyme in a mesophilic microorganism. However, the enzymatic activity of NspN-H was markedly reduced during purification, which hampered efforts to determine specific activity. Similar observations (a decrease in enzymatic activity during purification) were reported for other Gn-ATs.18, 19, 20 This decrease in activity was most likely due to oxidation of the free thiol of the catalytic cysteine (Cys-2). However, enzyme activity increased only slightly with increased concentrations of dithiothreitol (up to 5 mM) in the reaction buffer.
Stability of NspN. The stability of NspN is shown as a function of (a) pH and (b) heat. Britton–Robinson buffers (pH 4–11)21 were used to measure NspN activity using a standard assay (see Materials and methods) after storage at 16 °C for 18 h in solutions of various pH. For heat stability measurements, NspN was stored for 1 h at pH 7.5 at various temperatures and the NspN activity was measured as above.
Substrate specificity of NspN
The amino-acceptor substrate specificity of NspN was examined by in vitro NspN-H reactions using several carboxylic acid derivatives. The reaction products were identified by LC-ESIMS, and the enzyme activity was measured by a spectrophotometric method. The enzymatic activity of NspN-H toward various substrates is represented relative to that toward 3,4-AHBA (the most favorite substrate for NspN-H), which was defined as 100% (Figure 3). NspN-H has been shown to amidate 3,4-AHBA, but not 4,3-HNBA.11 Because 3,4-AHBA and 4,3-HNBA differ only by the functional group (-NH2 and –NO, respectively) at the meta position to the COOH group, benzoic acid derivatives with different functional groups at that position were evaluated as substrates for NspN-H. Amidation of protocatechuic acid (-OH), 4-hydroxy-3-methoxybenzoic acid (-OCH3) and p-hydroxybenzoic acid (-H) was catalyzed by NspN-H with relatively high efficiency (20, 32 and 39%, respectively). Thus, NspN disfavored only 4,3-HNBA among these m-substituted p-hydroxybenzoic acids. No activity was observed with 5-aminosalicylic acid and m-aminobenzoic acid, indicating the importance of the hydroxy group para to the carboxy group in m-aminobenzoic acids. No activity was also observed with 4-amino-3-hydroxybenzoic acid. In contrast, the amidation of benzoic acid derivatives that lacked an amino group was catalyzed by NspN-H with relatively high efficiency, regardless of the presence or position of hydroxy group(s). Examples include protocatechuic acid (20%), 4-hydroxy-3-methoxybenzoic acid (32%) and p-hydroxybenzoic acid (39%), all of which are described above, as well as 3-hydroxy-4-methylbenzoic acid (19%), m-hydroxybenzoic acid (8%), benzoic acid (7%) and salicylic acid (5%). Furthermore, NspN-H catalyzed the amidation of some carboxylate-type phenylpropanoids, trans-p-coumaric acid (13%), trans-cinnamic acid (7%) and trans-caffeic acid (6%), although the activities toward these substrates were generally lower than those for the corresponding benzoic acids, p-hydroxybenzoic acid (39%), benzoic acid (7%) and protocatechuic acid (20%), respectively. In contrast, L-aspartic-4-semialdehyde, a precursor of 3,4-AHBA, was not used as a substrate. This suggests that amidation by NspN occurs with 3,4-AHBA and not with its precursor, L-aspartic-4-semialdehyde, in the 4,3-HNBAm biosynthetic pathway.
Phylogenetic analysis of Gn-ATs
Protein databases were searched for bacterial Gn-ATs that seem to be involved in secondary metabolism processes, and a phylogenetic tree was constructed using four well-characterized asparagine synthases as the out-group (Supplementary Figure S1 and Supplementary Table S1). This analysis showed that NspN belonged to the same clade as several bacterial Gn-ATs, some of which had been evaluated for their function in secondary metabolite syntheses in vivo. Apart from the clade including NspN, several other bacterial Gn-ATs also seem to be involved in secondary metabolism processes, as suggested by the location of their genes in clusters generally associated with secondary metabolites. Asparagine synthetases, which catalyze the conversion of aspartic acid to asparagine, are phylogenetically distant from NspN.
Conclusions
NspN-H exhibited a broad amino-acceptor substrate specificity and catalyzed the amidation of (i) several m-substituted p-hydroxybenzoic acids except 4,3-HNBA, (ii) several benzoic acid derivatives with no amino group (a hydroxy group at the para position to the carboxy group was essential for m-aminobenzoic acids to be a substrate for NspN-H) and (iii) some carboxylate-type phenylpropanoids, such as trans-p-coumaric acid, trans-cinnamic acid and trans-caffeic acid. NspN-H showed the highest activity toward 3,4-AHBA among the several benzoic acid derivatives and carboxylate-type phenylpropanoids examined in this study. This result is reasonable because 3,4-AHBA is the natural substrate for NspN in 4,3-HNBAm biosynthesis. A phylogenetic analysis of Gn-ATs showed that NspN belongs to a clade of bacterial Gn-ATs that seem to be involved in secondary metabolism processes. NspN and related bacterial Gn-ATs may be useful in combinatorial biosyntheses of benzamide derivatives, which could be used as therapeutic agents for several diseases. This is the first report to describe the amino-acceptor substrate specificity of a bacterial Gn-AT involved in secondary metabolism.
References
Massiere, F. & Badet-Denisot, M. A. The mechanism of glutamine-dependent amidotransferases. Cell Mol. Life Sci. 54, 205–222 (1998).
Mouilleron, S. & Golinelli-Pimpaneau, B. Conformational changes in ammonia-channeling glutamine amidotransferases. Curr. Opin. Struct. Biol. 17, 653–664 (2007).
Schendel, F. J., Mueller, E., Stubbe, J., Shiau, A. & Smith, J. M. Formylglycinamide ribonucleotide synthetase from Escherichia coli: cloning, sequencing, overproduction, isolation, and characterization. Biochemistry 28, 2459–2471 (1989).
Mavrodi, D. V. et al. Functional analysis of genes for biosynthesis of pyocyanin and phenazine-1-carboxamide from Pseudomonas aeruginosa PAO1. J. Bacteriol. 183, 6454–6465 (2001).
Chin-A-Woeng, T. F., Thomas-Oates, J. E., Lugtenberg, B. J. & Bloemberg, G. V. Introduction of the phzH gene of Pseudomonas chlororaphis PCL1391 extends the range of biocontrol ability of phenazine-1-carboxylic acid-producing Pseudomonas spp. strains. Mol. Plant. Microbe. Interact. 14, 1006–1015 (2001).
Zhang, W., Ames, B. D., Tsai, S. C. & Tang, Y. Engineered biosynthesis of a novel amidated polyketide, using the malonamyl-specific initiation module from the oxytetracycline polyketide synthase. Appl. Environ. Microbiol. 72, 2573–2580 (2006).
Pickens, L. B. et al. Biochemical analysis of the biosynthetic pathway of an anticancer tetracycline SF2575. J. Am. Chem. Soc. 131, 17677–17689 (2009).
Seco, E. M., Miranzo, D., Nieto, C. & Malpartida, F. The pcsA gene from Streptomyces diastaticus var. 108 encodes a polyene carboxamide synthase with broad substrate specificity for polyene amides biosynthesis. Appl. Microbiol. Biotechnol. 85, 1797–1807 (2010).
Miranzo, D., Seco, E. M., Cuesta, T. & Malpartida, F. Isolation and characterization of pcsB, the gene for a polyene carboxamide synthase that tailors pimaricin into AB-400. Appl. Microbiol. Biotechnol. 85, 1809–1819 (2010).
Liao, R. et al. Thiopeptide biosynthesis featuring ribosomally synthesized precursor peptides and conserved posttranslational modifications. Chem. Biol. 16, 141–147 (2009).
Noguchi, A., Kitamura, T., Onaka, H., Horinouchi, S. & Ohnishi, Y . A copper-containing oxidase catalyzes C-nitrosation in nitrosobenzamide biosynthesis. Nat. Chem. Biol. 6, 641–643 (2010).
Maitre, M., Ratomponirina, C., Gobaille, S., Hode, Y. & Hechler, V. Displacement of [3H] gamma-hydroxybutyrate binding by benzamide neuroleptics and prochlorperazine but not by other antipsychotics. Eur. J. Pharmacol. 256, 211–214 (1994).
Terai, M., Usuda, S., Kuroiwa, I., Noshiro, O. & Maeno, H. Selective binding of YM-09151-2, a new potent neuroleptic, to D2-dopaminergic receptors. Jpn. J. Pharmacol. 33, 749–755 (1983).
Petrow, V. & Stephenson, O. Analgesics. III. Salicylamide derivatives. J. Pharm. Pharmacol. 10, 96–102 (1958).
Justin-Besancon, L. & Laville, C. Antiemetic action of metoclopramide with respect to apomorphine and hydergine. C. R. Seances. Soc. Biol. Fil. 158, 723–727 (1964).
Iwanaga, Y., Miyashita, N., Saito, T., Morikawa, K. & Itoh, Z. Gastroprokinetic effect of a new benzamide derivative itopride and its action mechanisms in conscious dogs. Jpn. J. Pharmacol. 71, 129–137 (1996).
Black, S. & Wright, N. G. Aspartic beta-semialdehyde dehydrogenase and aspartic beta-semialdehyde. J. Biol. Chem. 213, 39–50 (1955).
Patterson, M. K. Jr. & Orr, G. L-Asparagine biosynthesis by nutritional variants of the Jensen sarcoma. Biochem. Biophys. Res. Commun. 26, 228–233 (1967).
Milman, H. A. & Cooney, D. A. Partial purification and properties of L-asparagine synthetase from mouse pancreas. Biochem. J. 181, 51–59 (1979).
Hongo, S. & Sato, T. Some molecular properties of asparagine synthetase from rat liver. Biochim. Biophys. Acta. 742, 484–489 (1983).
Britton, H. T. S. & Robinson, R. A. Universal buffer solutions and the association constant of veronal. J. Chem. Soc. 1456–1462 (1931).
Acknowledgements
This work was supported, in part, by a research grant from the New Energy and Industrial Technology Development Organization of Japan, and by a Grant-in-Aid for Scientific Research (C) from the Ministry of Education, Culture, Sports, Science, and Technology of Japan (MEXT).
Author information
Authors and Affiliations
Corresponding author
Additional information
Dedicated to the late Dr C Richard Hutchinson for his exceptional contributions to natural product biosynthesis, engineering, and drug discovery.
Supplementary Information accompanies the paper on The Journal of Antibiotics website
Supplementary information
Rights and permissions
About this article
Cite this article
Noguchi, A., Horinouchi, S. & Ohnishi, Y. Substrate specificity of benzamide synthetase involved in 4-hydroxy-3-nitrosobenzamide biosynthesis. J Antibiot 64, 93–96 (2011). https://doi.org/10.1038/ja.2010.144
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ja.2010.144