Abstract
With the aim of identifying more novel natural epothilone derivatives produced by the epothilones A and B producing strain Sorangium cellulosum strain So0157-2, a large-scale fermentation (5000 l) of the strain was carried out. As a result, five new epothilone variants (1–5) were isolated from the fermentation broth. Their structures were established as 3-α-D-arabinofuranosides of epothilones A (1), B (2), D (3), C9 (4) and 8-demethyl epothilone A (5) by extensive NMR analysis and chemical methods. Bioassay results showed that compounds 1 and 2 had a weaker cytotoxic activity than did epothilone B.
Similar content being viewed by others
Introduction
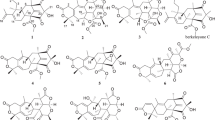
Epothilones are naturally occurring 16-membered ring macrolides that constitute a novel class of antimicrotubule-targeting agents. As the major products, epothilones A and B, were originally isolated from fermentations of the soil-derived myxobacterium Sorangium cellulosum So ce90,1, 2, 3 many natural epothilone analogs and related structures have been described.4, 5, 6 Moreover, synthetic and semisynthetic methods were important alternatives for obtaining epothilone analogs. At present, more epothilone variants are prepared in a synthetic manner.7, 8 Of all the epothilones and other variants coming from natural resources and synthesis, ixabepilones have been approved by FDA in 2007 for clinical use for the treatment of certain forms of breast cancer, in addition to a number of them being in preclinical and clinical trials.9 The attractive potential of epothilones led us to search for more potent and selective epothilone derivatives to satisfy the needs of chemotherapy for tumors. Presumably, when fermentation was scaled up, there were some minor and new analogs that were not isolated in small-scale fermentation. Hence, we investigated the chemical compositions of large-scale fermentation (5000 l) of epothilones A and B producing strain S. cellulosum strain So0157-2,10 and five new epothilone variants (1–5, Figures 1 and 2) were obtained. Their structures were established as 3-α-D-arabinofuranosides of epothilones A (1), B (2), D (3), C9 (4) and 8-demethyl epothilone A (5) by extensive NMR analysis and chemical methods. This paper describes the isolation and structure determination of the five new compounds and the cytotoxic activity of compounds 1 and 2.
Results and discussion
Structure elucidation
In the large-scale production and isolation of epothilones A and B from the strain S. cellulosum strain So0157-2, the byproducts, including other epothilones, were pooled and a crude extract was obtained. The pooling crude extract was isolated by silica gel column chromatography and semi-preparative HPLC to afford five new epothilone derivatives (1–5).
Compound 1 was isolated as a colorless oil. Its molecular formula of C31H47NO10S was deduced from high-resolution electrospray ionization mass spectrometry (HRESIMS) m/z 648.2840 ([M+Na]+, calcd 648.2818). The IR spectrum showed the hydroxyl absorption band at 3444 cm−1. The 1H NMR spectrum of 1 showed two characteristic downfield singlet signals and one singlet methyl signal of epopthilones A and B at δ 7.26, 6.55 and 2.69, respectively. In addition to one vinyllic methyl at δ 2.07, two aliphatic methyl doublets at δ 1.16 and 1.03 and two aliphatic methyl singlets at δ 1.20 and 1.32 were observed in the upfield. These data agreed well with those of epothilone A, except for a singlet proton signal at δ 5.12 and five proton signals between δ 4.04 and δ 3.60. The 13C NMR and distortionless enhancement by polarization transfer 135 (DEPT 135) spectra of 1 exhibited 32 carbons. Except for five oxygenated carbons (one acetal carbon, three oxygenated methines and one oxygenated methylene), the chemical shifts of the remaining carbons were almost identical to that of epothilone A. By a detailed analysis of 1H and 13C NMR data and 1H-1H correlation spectroscopy (1H-1H COSY) and heteronuclear multiple bond correlation (HMBC) spectra, a furanose was connected to the anomeric carbon at δ 109.0. These data showed that compound 1 included two moieties, aglycone was epothilone A and sugar was a furanose. The connection position of sugar to aglycone epothilone A was confirmed by HMBC experiments. The crossing peak between δ 5.12 (anomeric proton) and the δ 80.7 (C-3) indicated that sugar connected to epothilone A by an ether bond between C-3 and C-1′. Thus, the gross planar structure of 1 was established. The attempt to perform acid hydrolysis of compound 1 to obtain epothilone A was unsuccessful because of the instability of the three-membered ring between C-11 and C-12. By a detailed comparison of the 1H and 13C NMR data of 1 with those of epothilone A in conjunction with concurring with epothilone A, the relative and absolute configuration of the aglycone of 1 was assigned as that of epothilone A.
Acid hydrolysis of 1 afforded an aglycone and a sugar, and the sugar component identified by co-chromatography with authentic samples of arabinose, ribose and xylose on TLC analysis, and the identical RF values of the sugar moiety with those of arabinose indicated that the sugar moiety of 1 was arabinose. Absolute configuration for the arabinose was determined to be the D form according to the procedure developed by Tanaka et al.11 Comparison of the 13C NMR data of the furanose moiety of 1 with that of methyl α-D-arabinofuranoside and methyl β-D-arabinofuranoside reported in the literature,12 along with the small coupling constant (J=1.2 Hz) of the anomeric proton, assigned the configuration of the anomeric center (C-1′) to be α.
Compound 2 was obtained as a colorless oil. HRESIMS gave its molecular formula as C32H49NO10S. The 1H and 13C NMR data of 2 were similar to those of compound 1, except for the fact that a singlet methyl was present at δ 1.29 in the 1H NMR spectrum and an oxygenated quarternary carbon was present at δ 62.0 in the 13C NMR spectrum. Comparing the NMR data of the aglycone of 2 with those of epothilone B, the aglycone of 2 was very similar to epothilone B. Hence the aglycone of 2 was elucidated as epothilone B. The NMR data of sugar moiety were the same as those of 1 and were identified as α-D-arabinofuranose. The linkage of the sugar moiety to epothilone B was confirmed by the HMBC correlations of δ 5.09 (H-1′) and δ 78.6 (C-3).
Compound 3 was isolated as a colorless oil. The molecular formula of 3 was determined to be C31H47NO9S on the basis of HRESIMS analysis [m/z 632.2895 (M+Na)+] and was supported by 1H and 13C NMR data (Tables 1 and 2). The numbers of carbon of 3 were the same as that of 1. The only difference in NMR spectra between 3 and 1 was that the three-membered ring of C12-C13 in 1 was replaced by a double bond in 3. Comparing the NMR data of the aglycone of 3 with those of epothilone D, the aglycone of 3 was very similar to epothilone D and the aglycone of 3 was assigned to be epothilone D. The sugar moiety of 3 was established as α-D-arabinofuranose, similar to that of 1. The connection of the aglycone and sugar moiety was also supported by the HMBC correlation of δ 5.19 (H-1′) and δ 79.4 (C-3).
Compound 4 was assigned the molecular formula of C31H47NO10S, similar to that of 1, on the basis of HRESIMS analysis [m/z 648.2800 (M+Na)+] and NMR data (Tables 1 and 2). Analysis of the 1H and 13C NMR data for 4 revealed the presence of nearly identical structural features as those found in 3, except that the C-27 methyl in 3 was replaced by a hydroxylmethyl in 4. This result was supported by the 1H-13C long-range correlations between δH 6.61 (H-17) and δC 58.3 (C-27), 119.3 (C-19) and 79.6 (C-15), and between δH 4.35 (H-27) and δC 123.4 (C-17), 140.5 (C-16) and 79.6 (C-15). A comparison of the NMR data of the aglycone of 4 with those of epothilone C9 revealed the aglycone of 4 to be very similar to epothilone C9 and hence the aglycone of 4 was assigned to be epothilone C9. The sugar moiety of 4 was established as α-D-arabinofuranose, similar to that of 1. The connection of aglycone and sugar moiety was confirmed by the HMBC correlation of δ 5.18 (H-1′) and δ 78.8 (C-3).
Compound 5 has a molecular formula of C30H45NO10S, consistent with a structure having one less methyl group than does 1. Comparing the 1H NMR data of 5 with that of 1, only a doublet methyl was found to be absent in 5. The present HMBC correlation between δH 1.03 (H3-25) and δC 78.7 (C-7) in 1 was not observed in 5, and the 1H-1H COSY correlation of δ 3.85 (H-7) and a methylene signal at δ 1.46 and 1.60 (H2-8) suggested that the C-25 methyl in 1 was replaced by a hydrogen atom in 5. As a result, the aglycone of 5 was elucidated as 8-demethylepothilone A and the sugar moiety of 5 was established as α-D-arabinofuranose, similar to that of 1. The connection of aglycone of 5 and sugar moiety was supported by the HMBC correlation of δ 5.15 (H-1′) and δ 78.5 (C-3). Thus, the structure of 5 was established.
Biological activity
Compounds 1 and 2 were evaluated for cytotoxic activity against two tumor cell lines by CCK-8 methods,13 and the results showed that compounds 1 and 2 had a weaker cytotoxic activity than did epothilone B (Table 3).
Previous studies with regard to the structure–activity–relationship of various epothilone analogs had established that, in general, changes in the 16-membered macrolide framework led to a loss of tubulin affinity. For instance, sterechemical inversion14 and ring contraction/expansion15 reduced tubulin-binding affinity. Specifically, the C-3 diastereomer (3R) of epothilone A had been reported to be inactive in both tubulin polymerization and cytotoxicity assays.16 A recent study showed that the 3-hydroxyl group may not be directly involved in the fundamental network of interactions between epothilones and b-tubulin.17 This suggested that the less potent of 1 and 2, than epothilone B, may be attributed to the change of conformation of the 16-membered macrolide framework influenced by the 3-α-D-arabinofuranoside moiety. Further work should be carried out to clarify the conformation of 1 and 2 and the structure–activity–relationship of C-3-substituted epothilones. Moreover, compounds 1–5 demonstrate the possibility of structure–activity–relationship being involved in the development of the second generation of epothilone.
Experimental section
General experimental procedures
The UV spectra were obtained on a Varian CARY 300 BIO spectrophotometer (Varian Inc., Cary, NC, USA); IR spectra were recorded on a Nicolet Magna FT-IR 750 spectrometer (νmax in cm−1; Nicolet, Tokyo, Japan); 1H and 13C NMR spectra were measured with a Bruker DRX-400 (400 MHz for 1H and 100 MHz for 13C; Bruker, Rheinstetten, Germany) spectrometer. Chemical shifts are reported in parts per million (p.p.m.) (δ), using residual CHCl3 (δH 7.26 p.p.m.; δC 77.0) or CD3OD (δH 3.30 p.p.m.; δC 49.0) as an internal standard, with coupling constants (J) in Hz. 1H and 13C NMR assignments were supported by 1H-1H COSY, heteronuclear multiple quantum correlation and HMBC experiments. Electrospray ionization mass spectrometry (ESIMS) and HRESIMS spectra were taken on a Q-TOF Micro LC-MS-MS mass spectrometer (Agilent Technologies UK Ltd., Manchester, UK). The analytic (Zorbax SB-C18, 5 μm, 250 × 4.6 mm i.d.) and semi-preparative (Zorbax SB-C18, 5 μm, 250 × 9.4 mm i.d.) RP-HPLC was conducted on an Agilent 1100 series (Agilent Technologies Inc., Palo Alto, CA, USA). Commercial silica gel (Qing Dao Hai Yang Chemical Group Co., Qingdao, Shandong, China; 100–200 and 200–300 mesh) was used for column chromatography.
Myxobacterium material
The producing organism S. cellulosum strain So0157-2 was provided by Professor Yuezhong Li at the Shandong University, China and was deposited in the China Center of Typical Culture Collection (CCTCC) with accession no: CCTCC M 208078. Stock cultures were placed on a starch-yeast agar plate (0.8% soluble starch, 0.2% glucose, 0.2% peptone, 0.2% yeast extract, 0.2% MgSO4, 0.1% CaCl2, pH 7.8 autoclave at 121 °C for 30 min). Flask cultures were incubated at 30 °C on a rotary shaker at 250 r.p.m. for 4 days. Fermentation was carried out in a 50 l first seed fermentor (containing 30 l fermentation broth), in a 500 l second seed fermentor (containing 300 l fermentation broth), and finally in a 5000 l fermentor (containing 3000 l fermentation broth) successively. The following production medium was used: 1.0% soluble starch, 0.4% glucose, 0.2% peptone, 0.2% yeast extract, 0.2% MgSO4, 0.1% CaCl2, pH 7.8, autoclave. For the continuous adsorption of epothilones, 2.0% (v/v) of XAD-1600 (Rohm and Haas, Paris, France) was added. Fermentation was carried out at 28 °C for 12 days, stirring at 40 r.p.m. with an aeration rate of 120 m3 of air per hour.
Extraction and isolation
Adsorber resin (60 l) was separated from the 3000 l fermentation broth with a process filter. After washing the resin with water, the resin was eluted with four bed volumes of 95% ethanol to obtain the ethanol eluent. The ethanol eluent was diluted to about 30% ethanol and subjected to a XAD-1600 resin column, eluting with 30, 40, 50, 60 and 70% ethanol (each concentration eluted with two bed volumes). The eluents eluting with 60 and 70% ethanol were pooled and concentrated in vacuo at 50 °C to give a mixture. The mixture was dissolved in CHCl3 and chromatographed on a silica gel column eluting with petroleum ether/acetone from 70:30–40:60, and fractions 1–4 were obtained on the basis of the TLC profiles. Analysis of fraction 1 by HPLC showed that it mainly contained epothilones A and B. Fractions 2–4 were subjected to a Sephadex LH-20 column (Pharmacia, Uppsala, sweden) and eluted with ethanol and detected by TLC to give subfraction 2, subfraction 3 and subfraction 4, respectively. Subfraction 2 was further separated by semi-preparative HPLC eluting with MeOH/H2O (80:20) to give compound 3 (tR16.2 min, 21 mg). Subfraction 3 was also isolated by semi-preparative HPLC eluting with MeOH/CH3CN/H2O (35:10:55) to afford compounds 4 (tR 22.8 min, 11 mg) and 5 (tR 26.6 min, 8 mg). Subfraction 4 was further purified by semi-preparative HPLC eluting with CH3CN/H2O (45:55) to obtain compound 1 (tR 15.0 min, 55 mg) and one collection (tR 17.0 min). The collection was further separated by semi-preparative HPLC eluting with MeOH/CH3CN/H2O (35:10:55) to give compound 2 (tR 28.1 min, 10 mg).
Epothilone A 3-α-D-arabinofuranoside (1)
Colorless oil; [α]25 D-13.3 (c 0.3, EtOH); UV (EtOH) λmax 249 (ɛ 12083), 206 (ɛ 20416) nm; IR (KBr): vmax 3444, 1738, 1689 cm−1; 1H and 13C NMR data, see Tables 1 and 2; ESIMS m/z 626 [M+H]+, 648 [M+Na]+; HRESIMS m/z 648.2840 [M+Na]+ (calcd for C31H47NO10SNa, 648.2818).
Epothilone B 3-α-D-arabinofuranoside (2)
Colorless oil; [α]25 D-3.57 (c 0.28, EtOH); UV (EtOH) λmax 249 (ɛ 10437), 212 (ɛ 13845) nm; IR (KBr): vmax 3455, 1739, 1689 cm−1; 1H and 13C NMR data, see Tables 1 and 2; ESIMS m/z 640 [M+H]+, 662 [M+Na]+; HRESIMS m/z 662.2982 [M+Na]+ (calcd for C32H49NO10SNa, 662.2975).
Epothilone D 3-α-D-arabinofuranoside (3)
Colorless oil; [α]25 D-27.4 (c 0.84, EtOH); UV (EtOH) λmax 251 (ɛ 16443), 213 (ɛ 20503) nm; IR (KBr): vmax 3457, 1738, 1687 cm−1; 1H and 13C NMR data, see Tables 1 and 2; ESIMS m/z 610 [M+H]+, 632 [M+Na]+; HRESIMS m/z 632.2895 [M+Na]+ (calcd for C31H47NO9SNa, 632.2869).
Epothilone C9 3-α-D-arabinofuranoside (4)
Colorless oil; [α]25 D-26.2 (c 0.42, EtOH); UV (EtOH) λmax 254 (ɛ 10714), 209 (ɛ 9970) nm; IR (KBr): vmax 3446, 1737, 1688 cm−1; 1H and 13C NMR data, see Tables 1 and 2; ESIMS m/z 626 [M+H]+, 648 [M+Na]+; HRESIMS m/z 648.2800 [M+Na]+ (calcd for C31H47NO10SNa, 648.2818).
8-Demethyl epothilone A 3-α-D-arabinofuranoside (5)
Colorless oil; [α]25 D+7.9 (c 1.13, EtOH); UV (EtOH) λmax 250 (ɛ 7087), 214 (ɛ 8798) nm; IR (KBr): vmax 3438, 1736, 1690 cm−1; 1H and 13C NMR data, see Tables 1 and 2; ESIMS m/z 612 [M+H]+, 634 [M+Na]+; HRESIMS m/z 634.2681 [M+Na]+ (calcd for C30H45NO10SNa, 634.2662).
Acid hydrolysis of 1
Compound 1 (4.4 mg). was dissolved in 1 M HCl (1 ml) and then heated at 80 °C for 4 h. Aglycone was extracted with CHCl3 thrice, and the aqueous residue was evaporated under reduced pressure. The residue was compared with standard sugars of arabinose, ribose and xylose (Sigma, St Louis, MO, USA) using TLC [n-butanol: CH3COOH: H2O=4:1:1] analysis, which showed the sugar (RF=0.4) to be arabinose.
Determination of the absolute configuration of arabinose
The above drying aqueous residue was dissolved in pyridine (0.1 ml) containing L-cysteine methyl ester hydrochloride (1 mg) and heated at 60 °C for 1 h. Arylisothiocyanate (1.2 μl) was added to the mixture, which was heated at 60 °C for 1 h. The reaction mixture was directly analyzed by reversed-phase HPLC on an ODS column (4.6 × 250 mm) at 35 °C with an isocratic elution of 25% CH3CN in 50 mM H3PO4 for 30 min and a subsequent washing of the column with 90% CH3CN at a flow rate of 0.8 ml min−1. Peaks were detected with an ultraviolet detector at 250 nm. The peak of the reaction mixture was detected at 15.72 min (D-arabinose). Treated in the same manner, standard D-arabinose (Sigma) gave a peak at 15.76 min and L-arabinose (Sigma) gave a peak at 14.60 min.
Antitumor bioassays
For cytotoxicity measurements, stock epothilonoside solutions were prepared at 100 μg/ml in dimethyl sulfoxide and stored at −20 °C. Human lung carcinoma A549, large intestine cancer cells LOVO and Human Ovarian Cancer Cell Line SKOV-3 were obtained from the Institute of Biochemistry and Cell Biology, Shanghai, Chinese Academy of Sciences. All cell lines were routinely cultured in Dulbecco’s modified Eagle’s medium containing 10% calf serum at 37 °C for 4 h, in a humidified atmosphere of 5% CO2 incubator. The adherent cells at their logarithmic growth stage were digested, and were inoculated onto a 96-well culture plate at a density of 1.0 × 104/well for the determination of proliferation. Test samples ranging from 0.1–100 μg ml−1 in 100 μl were added to cells in triplicate wells, and incubation was continued for 72 h. Coloration substrate, cell counting kit-8 (CCK-8, Dojindo, Kumamoto, Japan), was added to the medium, followed by further incubation for 3 h. Absorbance at 450 nm with a 600 nm reference was measured thereafter. Media and dimethyl sulfoxide control wells, in which compound was absent, were included in all the experiments in order to eliminate the influence of dimethyl sulfoxide. The inhibitory rate of cell proliferation was calculated by the following formula:
Growth inhibition (%)=[ODcontrol−ODtreated]/ODcontrol × 100%.
The cytotoxicity of compound on tumor cells was expressed as IC50 values (the drug concentration reducing the absorbance in treated cells by 50%, with respect to untreated cells) and was calculated by the LOGIT method.
1H and 13C NMR spectra of five new epothilone variants (1–5) are detailed in Supplementary information.
Accession codes
References
Höfle, G., Bedorf, N., Gerth, K. & Reichenbach, H. (GBF). Epothilones, process for preparing the same and their use as medicaments and as plant protecting agents. D.E. 4,138,042, May 27 (1993).
Gerth, K., Bedorf, N., Höfle, G., Irschik, H. & Reichenbach, H. Epothilons A and B: antifungal and cytotoxic compounds from Sorangium cellulosum (Myxobacteria). production, physico-chemical and biological properties. J. Antibiot. 49, 560–563 (1996).
Höfle, G. et al. Epothilone A and B-novel 16-membered macrolides with cytotoxic activity: isolation, crystal structures, and conformation in solution. Angew. Chem. Int. Ed. Engl. 35, 1567–1569 (1996).
Hardt, I. H. et al. New natural epothilones from Sorangium cellulosum, strains So ce90/B2 and So ce90/D13: isolation, structure elucidation, and SAR studies. J. Nat. Prod. 64, 847–856 (2001).
Arslanian, R. L. et al. A new cytotoxic epothilone from modified polyketide synthases heterologously expressed in Myxococcus xanthus. J. Nat. Prod. 65, 1061–1064 (2002).
Starks, C. M., Zhou, Y., Liu, F. & Licari, P. J. Isolation and characterization of new epothilone analogues from recombinant Myxococcus xanthus fermentations. J. Nat. Prod. 66, 1313–1317 (2003).
Nicolaou, K. C., Roschangar, F. & Vourloumis, D. Chemical biology of epothilones. Angew. Chem. Int. Ed. Engl. 37, 2014–2045 (1998).
Dong, S. D. et al. Rapid access to epothilone analogs via semisynthetic degradation and reconstruction of epothilone D. Tetrahedron Lett. 45, 1945–1947 (2004).
Lee, J. J. & Swain, S. M. The epothilones: translating from the laboratory to the clinic. Clin. Cancer Res. 14, 1618–1624 (2008).
Gong, G. L. et al. Mutation and a high-throughput screening method for improving the production of epothilones of Sorangium. J. Ind. Microbiol. Biotechnol. 34, 615–623 (2007).
Tanaka, T., Nakashima, T., Ueda, T., Tomii, K. & Kouno, I. Facile discrimination of aldose enantiomers by reversed-phase HPLC. Chem. Pharm. Bull. 55, 899–901 (2007).
Gorin, P. A. J. & Mazurek, M. Further studies on the assignment of signals in 13C magnetic resonance spectra of aldoses and derived methyl glycosides. Can. J. Chem. 53, 1212–1223 (1975).
Wang, J. D. et al. HS071, A new furan-type cytotoxic metabolite from Streptomyces sp. HS-HY-071. J. Antibiot. 61, 623–626 (2008).
Nicolaou, K. C. et al. Designed epothilones: solid phase synthesis on microtubes, tubulin assembly properties and cytotoxic action against taxol-resistant tumor cells. Angew. Chem. Int. Ed. Engl. 36, 2097–2103 (1997).
Nicolaou, K. C. et al. Probing the ring size of epothilones: total synthesis of [14]-, [15]-, [17]-, and [18]-epothilones A. Angew. Chem. Int. Ed. Engl. 37, 81–84 (1998).
Meng, D. et al. Remote effects in macrolide formation through ring-forming olefin metathesis: an application to the synthesis of fully active epothilone congeners. J. Am. Chem. Soc. 119, 2733–2734 (1997).
Erdélyi, M. et al. Conformational preferences of natural and C3-modified epothilones in aqueous solution. J. Med. Chem. 51, 1469–1473 (2008).
Author information
Authors and Affiliations
Corresponding author
Additional information
Supplementary Information accompanies the paper on The Journal of Antibiotics website (http://www.nature.com/ja)
Rights and permissions
About this article
Cite this article
Wang, J., Zhang, H., Ying, L. et al. Five new epothilone metabolites from Sorangium cellulosum strain So0157-2. J Antibiot 62, 483–487 (2009). https://doi.org/10.1038/ja.2009.55
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ja.2009.55
Keywords
This article is cited by
-
Micromonospora zhangzhouensis sp. nov., a Novel Actinobacterium Isolated from Mangrove Soil, Exerts a Cytotoxic Activity in vitro
Scientific Reports (2020)
-
Two new glutarimide antibiotics from Streptomyces sp. HS-NF-780
The Journal of Antibiotics (2019)
-
Two new lankacidin-related metabolites from Streptomyces sp. HS-NF-1178
The Journal of Antibiotics (2018)
-
Two new spliceostatin analogs from the strain Pseudomonas sp. HS-NF-1408
The Journal of Antibiotics (2018)
-
A new anthracycline-type metabolite from Streptomyces sp. NEAU-L3
The Journal of Antibiotics (2017)