Abstract
Marine and lake snow is a continuous shower of mixed organic and inorganic aggregates falling from the upper water where primary production is substantial. These pelagic aggregates provide a niche for microbes that can exploit these physical structures and resources for growth, thus are local hot spots for microbial activity. However, processes underlying their formation remain unknown. Here, we investigated the role of chemical signaling between two co-occurring bacteria that each make up more than 10% of the community in iron-rich lakes aggregates (iron snow). The filamentous iron-oxidizing Acidithrix strain showed increased rates of Fe(II) oxidation when incubated with cell-free supernatant of the heterotrophic iron-reducing Acidiphilium strain. Amendment of Acidithrix supernatant to motile cells of Acidiphilium triggered formation of cell aggregates displaying similar morphology to those of iron snow. Comparative metabolomics enabled the identification of the aggregation-inducing signal, 2-phenethylamine, which also induced faster growth of Acidiphilium. We propose a model that shows rapid iron snow formation, and ultimately energy transfer from the photic zone to deeper water layers, is controlled via a chemically mediated interplay.
Similar content being viewed by others
Introduction
Pelagic aggregates form in the water column through adsorption of inorganic and organic matter, including bacteria, phytoplankton, feces, detritus and bio-minerals (Alldredge and Silver, 1988; Simon et al., 2002; Thornton, 2002). These aggregates, also called marine or lake snow, typically range in size from millimeters to centimeters (Alldredge and Silver, 1988; Passow et al., 2012) and are held together by extracellular polysaccharides (Thornton, 2002; Giani et al., 2005; Passow et al., 2012). Marine and lake snow contribute substantially to the energy transfer from the photic zone to deeper water layers (Suess, 1980). During the passage to the sediments, many motile bacteria and zooplankton colonize these organic-rich particles and remain attached as the aggregates sink through the water column to the sediment (Fenchel, 2001; Kiørboe et al., 2003). Particle-associated bacteria show higher extracellular enzymatic activities than free-living, planktonic microbes, resulting in rapid and localized turnover of organic matter and its subsequent release into the surrounding water (Grossart and Simon, 1998). Pelagic aggregates are local hot spots for microbial interactions by direct cell contact, feeding activity and also the action of diffusible signals. For example, α-Proteobacteria, including Roseobacter strains, growing on the surface of marine snow produce elevated amounts of inhibitory molecules compared with planktonic forms in the surrounding aquatic environment (Long and Azam, 2001). N-acyl homoserine lactones, compounds known to function as potential quorum-sensing mediators, are produced by Roseobacter strains (Gram et al., 2002; Zan et al., 2014) and Pantoea sp. (Jatt et al., 2015) in marine snow communities, suggesting that quorum-sensing mechanisms might be at play. Chemical signaling molecules have also been suspected to regulate bacterial colonization, coordinate group behavior, as well as antagonistic activities within pelagic aggregates (Dang and Lovell, 2016). However, the exact mechanisms for chemical communication and the identity of the compounds mediating such interactions in pelagic aggregates are poorly understood.
The chemical diversity of exuded metabolites within such aggregates makes the identification and elucidation of infochemicals highly challenging, due to the loss of the diluted signals during the purification process (Prince and Pohnert, 2010). However, comparative metabolomics has emerged as a powerful tool to overcome the limitations of bioassay-guided approaches by allowing the identification of metabolites produced by microorganisms grown in co-culture by comparison with those of single-strain cultures. Notably, upregulated metabolites are prime-candidates for mediators of microbial interactions (Gillard et al., 2013; Kuhlisch and Pohnert, 2015).
In this study, we isolated two predominant bacterial key players from iron-rich lake aggregates (iron snow) to investigate chemical communication mechanisms involved in their interactions. Iron snow is characterized by lower microbial and chemical complexity in comparison with typical organic-rich marine and lake snow aggregates, and shows high sinking velocities because of its high relative fraction of iron (Reiche et al., 2011). We report that an active control by chemical signals shapes the association of the dominant microbial consortia within the iron snow and their behavior. Comparative metabolomics approach and structure elucidation led to the identification of bacterial extracellular exudates that function as allelopathic aggregate-inducing signal. These results clearly show that even during the very limited time of passage through the water column, interspecies chemical interactions between key organisms in iron snow enable these microorganisms to adhere to and colonize the particles.
Materials and methods
Sampling, bacterial isolation and identification
For bacterial isolation from iron snow, lake water was collected in the water column of the central basin (pH 2.8–3.0) and the northern basin (pH 5.4–5.6) just below the redoxcline of the acidic lignite mine lake (Lake 77) located in the Lusatian mining area in Germany, as described in a previous report (Mori et al., 2016). Sampling was carried out in September 2012, January and November 2013. The lake water was collected and stored at 4 °C and serially diluted (10−1 to 10−3) with filter-sterilized lake water and 100 μl of the dilution was plated onto three types double-layer media (Johnson and Hallberg, 2007; Lu et al., 2013; pH 2.5–5.5): iFeo, Feo and YEo that contain 25 mM ferrous sulfate and 0.25% tryptone soya broth (Feo) or 0.2% yeast extract (YEo), or without carbon source (iFeo). Cycloheximide (Carl Roth, Karlsruhe, Germany) was added into the top-layer medium at 50 μg ml−1 to inhibit the growth of Fungi. Bacterial growth was checked during incubation at room temperature in the dark. Colonies appeared were transferred into new media at least five times until pure isolates were obtained. Their purity was evaluated by microscopic cell observations and bacterial 16S rRNA gene sequencing.
Phylogenetic analyses and identification of the isolates were carried out by genomic DNA extraction and 16S rRNA gene sequencing. Cell colonies were suspended into 20 μl of lysis solution containing 0.05 M NaOH and 0.25% SDS and heated for 15 min at 95 °C, followed by centrifugation at 4000 g for 5 min to obtain genomic DNA in the supernatant. A 1 μl of the supernatant was subjected to PCR employing primer set 27F/1492R (Lane, 1991). The PCR product was purified through a spin column (NucleoSpin Gel and PCR Clean-up, Macherey-Nagel, Düren, Germany) and sequenced (Macrogen Europe, Amsterdam, The Netherlands). Raw sequences were processed in Geneious 4.6.1 for trimming and assembling, followed by BLAST homology search (Johnson et al., 2008). Sequences of the representative isolates were submitted to GenBank database under accession numbers LN866581 to LN866603 (Supplementary Table S1).
Growth tests of selected bacterial isolates
Selected bacterial isolates Acidithrix strain C25 and Acidiphilium strain C61 from the central basin were transferred into different types of liquid media (Supplementary Table S2), and their growth was tested to figure out an appropriate medium suitable for co-cultivating different bacterial species. The artificial pilot-plant water (APPW) medium (pH 2.5, 25 mM FeSO4, 0.022 g l−1 Na2SO4, 0.024 g l−1 K2SO4, 3.24 g l−1 MgSO4·7H2O, 0.515 g l−1 CaSO4·2H2O, 0.058 g l−1 NaHCO3, 0.010 g l−1 NH4Cl, 0.014 g l−1 Al2(SO4)3·18H2O, 0.023 g l−1 MnCl2·4H2O, 0.0004 g l−1 ZnCl2) reported by Tischler et al. (2013) with additional yeast extract (0.2 g l−1) was tested in addition to iFeo, Feo and YEo.
Monitoring of cell growth and morphology
Bacterial growth was confirmed by microscope, growth of Fe(II)-oxidizing bacteria was also determined by checking rust-colored Fe(III) oxide precipitates in the culture and measuring concentrations of Fe(II) and total dissolved Fe using the phenanthroline method (Tamura et al., 1974). Bacterial cell morphology was observed under a fluorescence microscope (Axioplan, Zeiss, Oberkochen, Germany). Cells collected from the culture were stained with SYTO 13 nucleic acid staining (Thermo Scientific, Waltham, MA, USA) on glass slides and visualized. The fluorescent images were recorded by a mono color camera (Axio Cam MRm, Zeiss). Total nucleic acids extraction from bacterial cells and subsequent quantification were conducted to estimate bacterial biomass in cultures. Bacterial cells were collected through centrifugation at 4000 g for 5 min, and subjected to bead beating in sodium phosphate buffer (pH 8.0) with TNS solution (500 mM Tris-HCl pH 8.0, 100 mM NaCl, 10% SDS wt vol−1). The supernatant was removed after centrifugation. Next, extraction with equal volumes of phenol-chloroform-isoamyl alcohol [25:24:1 (v/v/v)] and chloroform-isoamyl alcohol [24:1 (v/v)] was performed. Nucleic acids were precipitated with two volumes of polyethylene glycol 6000 (Carl Roth) solution by centrifugation at 20 000 g at 4 °C for 90 min. The pellets were washed with ice-cold 70% ethanol and suspended in 50 μl elution buffer (Qiagen, Hilden, Germany), followed by quantification using NanoDrop 1000 spectrophotometer (Thermo Scientific). As a more sophisticated technique to determine bacterial biomass, quantitative PCR (qPCR) analysis using primer set Uni388F-RC/Uni-907R (Lane, 1991) was conducted to monitor the bacterial 16S rRNA gene copy number. Aliquots of 1.25 ng DNA extracts were used in triplicate as template for qPCR on a Mx3000P real-time PCR system (Agilent, Santa Clara, CA, USA) with Maxima SYBR Green qPCR Mastermix (Thermo Scientific). Standard curves were prepared by serial dilution of plasmid DNA that contains the cloned 16S rRNA gene sequence of an uncultured bacterium (GenBank accession no. HE604015). Melting curve analysis was performed to confirm the specificities of the qPCR products.
Supernatant exchange experiment
Acidithrix strain C25 and Acidiphilium strain C61 cultures were grown in a 200 ml APPW+yeast extract (0.2 g l−1) medium for 3–7 days with shaking at room temperature, until Acidithrix cultures began to oxidize Fe(II). After 3–7 days of growth, the cultures were centrifuged and passed through a 0.22-μm filter and the cell-free supernatant was collected. A volume of 100 ml of cell-free supernatant was then added to 100 ml of another set of cultures (50% v/v), such that the cell-free supernatant obtained from the Acidithrix culture was added to cultures containing either Acidithrix or Acidiphilium and cell-free supernatant obtained from the Acidiphilium culture was added to cultures containing either Acidithrix or Acidiphilium (Figure 1a). In order to achieve sufficient statistical power, all experiments were conducted as biological triplicate. Bacterial Fe(II) oxidation was monitored every 1–3 days. Bacterial cells were stained using SYTO 13 and cell morphology was monitored using fluorescent microscopy.
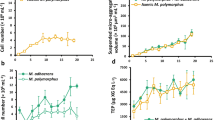
Supernatant exchange experimental setup, dissolved Fe(II)/total Fe concentrations over time and photographs of selected cultures. (a) Bacterial culture setup. Pure cultures of Acidithrix strain C25 (Acx) and Acidiphilium strain C61 (Acp) were amended with cell-free supernatants obtained from either same or different bacterial strain. (b, c) Concentrations of dissolved Fe(II) (b) and total Fe (c) in the cultures. Faster consumption of Fe(II) (P<0.001, Welch’s t-test) and total dissolved Fe (P<0.01) were observed in Acx (Acp) cultures compared with Acx (Acx) between days 1–4 and days 2–4, respectively (highlighted in gray). *Concentrations at time point zero were not measured; here the initial concentration of 25 mM is plotted. n=3, error bars indicate standard deviation. (d) Photographs depicting the appearances of Acidithrix strain C25 cultures amended with Acidithrix strain C25 supernatant [Acx (Acx), upper] or Acidiphilium strain C61 supernatant [Acx (Acp), lower]. Photos were taken at days 1, 3 and 7 of incubation and show relatively faster precipitation of rust colored Fe(III) precipitates in Acx (Acp) cultures compared with Acx (Acx) cultures.
Metabolomics methods
Sample preparation
A volume of 50 ml of the respective liquid cultures was passed through 0.22-μm filters after centrifugation at 4000 g. Solid-phase extraction (Oasis HLB 30 mg, 1-ml cartridge, Waters, Milford, MA, USA) of the filtrate was carried out using a 50-ml syringe without plunger, attached with the luer cone to a tapered solid-phase extraction adaptor that was placed onto the cartridge. Conditioning was carried out by passing 1 ml methanol followed by 1 ml ultra high performance liquid chromatography-grade water. After 50 ml sample was passed through the cartridge, 1 ml water was used to remove salts followed by air drying for 5 min. Elution was carried out with 1 ml of a mixture of methanol and acetonitrile 1:1 (v/v) by gravity. The samples were spiked with internal standard (5 μl of a 40 μM ribitol dissolved in water) and dried in a nitrogen stream followed by drying overnight in fine vacuum. The samples were dissolved in pyridine (20 μl) and treated with N,O-bis(trimethylsilyl)trifluoroacetamide BSTFA (20 μl) vortexed for 5 sec and kept at 60 °C for 1 h. GC/MS analyses were measured subsequently.
GC/MS measurement
Gas-chromatographic separations were executed on a Trace 1310 (Thermo Scientific) equipped with TriPlus RSH auto sampler (Thermo Scientific) and coupled with a TSQ 8000 electron impact (EI) triple quadrupole mass spectrometer (Thermo Scientific). We used a TG-5SILMS column (Thermo Scientific) with the following dimensions: length 30 m; 0.25-mm inner diameter and 0.25-μm film. The column was operated with helium carrier gas using a PTV injector operating with a column flow 1.2 ml min−1 and a split less injection for 1 min. After initial 60 °C the temperature was raised to 320 °C at a rate of 14.5 °C min−1 and then held for 2 min. For cleaning, the temperature was increased to 350 °C and held for 5 min with a flow of 50 ml min−1. After split less time, the split flow was set to 20 ml min−1. The syringe was cleaned twice with 5 μl n-hexane each before injection and rinsed with 1 μl sample before the injection was done. After injection, the syringe was washed five times with ethyl acetate and five times with n-hexane (5 μl each). The GC oven program started at 100 °C for 1 min and the temperature was increased to 320 °C at 5 °C min−1, and held for 3 min. Measurement using the mass spectrometer was begun after 10 min of monitoring. The mass range between 50 and 650 m/z were monitored. The MS transfer line was set to 300 °C as well as the ion source temperature.
XCMS analysis
The XCMS data processing was carried out with cdf files, which were converted from the Thermo RAW-files with the Xcalibur 3.0.63 (Thermo Scientific) on board file converter. Processing was carried out using XCMS Server version 3.01.01. We used the pre-defined settings for GC-measurement ‘Single Quad (matched filter)’; additionally the retention time correction was removed (Tautenhahn et al., 2012). The parameters used are listed in Supplementary Table S3.
Ultra high performance liquid chromatography/high resolution mass spectrometry measurement
To an aliquot of 100 μl bacterial culture, 100 μl methanol was added and the mixture was vortexed for 1 min in a 1.5-ml Eppendorf tube. The mixture was then centrifuged at 10 000 g. The supernatant was filled into a standard 1.5-ml HPLC vial with insert and sealed with a polytetrafluoroethylene-coated rubber seal. For each sample 10 μl were injected three times in a randomized sample list. Ultra high performance liquid chromatography coupled with high resolution mass spectrometry was carried out using a UltiMate HPG-3400 RS binary pump (Thermo Scientific), WPS-3000 auto sampler (Thermo Scientific) equipped with a 25-μl injection syringe and a 100-μl sample loop. The column was kept at 25 °C within the column compartment TCC-3200. The compounds were detected using a Kinetex C-18 RP chromatography column (50 × 2.1 mm; 1.7 μm; Phenomenex, Torrance, CA, USA) using the gradient specified in the Supplementary Table S4. Eluent A was water, with 2% acetonitrile and 0.1% formic acid. Eluent B was pure acetonitrile. Mass spectra were recorded with QExactive plus orbitrap mass spectrometer (Thermo Scientific). For monitoring, phenylalanine-targeted selective ion monitoring in the positive ionization mode was used with the following parameters: target ion [M+H]+ (m/z=122.1±1.0); resolution: 70 000; AGC target: 5 × 104; maximum IT: 200 ms; sheath gas flow rate: 60; aux gas flow rate: 20; sweep gas flow rate: 5; spray voltage: 3.3 kV; capillary temperature: 360 °C; S-lens RF level: 50; aux gas heater temperature: 400 °C; acquisition time frame: 0.5–2.5 min.
Quantification
Peak detection and integration was carried out using TRACEFINDER EFS 3.1 (Thermo Scientific) with the following settings: mass tolerance: 3 ppm; mass precision: 4; compound: C8H11N; adduct: [M+H]+; retention time window: 1.40 min (40 s); signal: XIC from SIM experiment; peak detection algorithm: genesis; peak detection method: nearest RT (Smoothing=1). Each technical sample was measured two times using a randomized sample list. The calibration standards were prepared from a 1 mM 2-phenethylamine (PEA) stock solution in the medium as matrix. Starting from this solution the working solution with a concentration 100 μM was prepared. From this working solution, calibration standards with a concentration of 5, 10, 15, 20, 25, 50, 75 and 100 μM, respectively, were prepared. The calibration curve was calculated as shown in Supplementary Figure S1.
Screening of allelochemicals
To evaluate the effects of the chemicals detected through the metabolomics profiling, commercial grade pure PEA (99%, Alfa Aesar, Ward Hill, MA, USA), p-hydroxybenzoic acid (99%, Alfa Aesar) and furane 2,5-dicarboxylic acid (98%, Alfa Aesar) were supplied to the pure bacterial cultures to reach final concentrations of 1–1000 μM. Cell growths and morphologies were monitored during incubations.
Results
Bacterial isolation and cultivation
In total, we obtained 133 bacterial isolates on different media designed to cultivate acidophilic bacteria from iron snow sampled below the redoxcline of the Lausatian acidic mining lake 77, Germany. Screened isolates were phylogenetically identified as either acidophilic Fe(II)-oxidizing bacteria related to Acidithrix (Actinobacteria) or Ferrovum (β-Proteobacteria), or as acidophilic heterotrophic Fe(III)-reducing bacteria related to Acidiphilium, Acidisphaera and Acidocella (α-Proteobacteria; Supplementary Table S1). These genera of Fe(II) oxidizers and Fe(III) reducers were previously reported to cover more than 50% of total microbial community in iron snow samples (Lu et al., 2013). We selected one key player from each group with more than 10% relative abundance for further in-depth investigation. The Fe(II)-oxidizing Acidithrix strain C25 (LN866582) was recently shown to form long filaments during early growth followed by large cell-mineral aggregates in late stationary phase (Jones and Johnson, 2015; Mori et al., 2016). The Fe(III)-reducing Acidiphilium strain C61 (LN866588) is a facultative anaerobe that can completely oxidize many carbon compounds including various sugars and organic acids (Küsel et al., 1999). For co-cultivation and supernatant exchange experiments, we identified a medium that allows identical growth conditions of these two key players. Both isolates could be grown under oxic conditions in APPW (Tischler et al., 2013) with additional yeast extract as nutrient source for the heterotrophic growth.
Supernatant exchange experiment
Since co-culture devices that allow spatial separation of the single cultures while maintaining the exchange of dissolved chemicals via diffusion (Paul et al., 2013) are not applicable for bacteria forming solid-state products that inhibit diffusion via a membrane, we performed a supernatant exchange experiment in order to detect extracellular chemical mediators produced by our isolates (Figure 1a). Cultures of Acidithrix and Acidiphilium were passed through a 0.22-μm filter membrane to obtain cell-free supernatants. Another set of Acidithrix and Acidiphilium cultures were amended with these cell-free supernatants. Four combinations were incubated for the co-culture experiments [Acx (Acx): Acidithrix cultures with Acidithrix supernatant; Acx (Acp): Acidithrix cultures with Acidiphilium supernatant; Acp (Acx): Acidiphilium cultures with Acidithrix supernatant; Acp (Acp): Acidiphilium cultures with Acidiphilium supernatant] (Figure 1a).
The addition of cell-free Acidiphilium supernatant to cultures of Acidithrix [Acx (Acp)] resulted in the oxidation of Fe(II) at a rate approximately 1.7-times faster (5.72 mM day−1) during days 1–4 of incubation than in the control cultures Acx (Acx) (3.39 mM day−1; P<0.001; Figure 1b). The Fe(II) oxidation rates in Acx (Acx) were comparable to those in pure Acidithrix cultures without amendment reported in a previous study (Mori et al., 2016). In addition, precipitation of insoluble Fe(III) oxides was 2.2-times faster (P<0.01) in these cultures during days 2–4 of incubation (Figures 1c and d). Total nucleic acid extracts yielded significantly higher amounts with 0.45 ng μl−1 in Acx (Acp) cultures after 10 days of incubation than in Acx (Acx) cultures (0.34 ng μl−1; P<0.01). No measurable abiotic Fe(II) oxidation was observed in cultures containing Acidiphilium and the cell-free controls.
Pronounced morphological changes were observed when cell-free supernatant of Acidithrix was added to Acidiphilium cultures. Macroscopic cell aggregates were formed in these Acp (Acx) cultures after 5–6 days of growth (Figures 2a2 and b2) in contrast to the Acp (Acp) cultures, which showed characteristic single, planktonic cells and small populations of aggregates (Figures 2a1 and b1).
Acidiphilium strain C61 cultures amended with cell-free supernatants of Acidiphilium strain C61 or Acidithrix strain C25. Photographs (a) and microscopic images (b) of Acidiphilium strain C61 cultures amended with cell-free supernatant of Acidiphilium strain C61 [Acp (Acp), a1 and b1] or of Acidithrix strain C25 [Acp (Acx), a2 and b2] taken at day 10 of incubation. The bacterial cells were stained with SYTO 13, a nucleic acid stain. Black arrow points to the macroscopic cell aggregates precipitated in the cultures.
Profiling of bacterial extracellular products
The induction of morphological changes as well as the alteration of metabolic processes by amendment of the cell-free supernatants clearly indicated the action of chemical interaction mediators. To gain deeper insight into the chemical communication between Acidithrix and Acidiphilium, we subjected the cell-free supernatants (Figure 1a) to solid-phase extraction after a 10-day incubation period. The extracts were derivatized through silylation and subsequently measured with GC/MS. The m/z-retention time pairs were extracted from the chromatograms and processed using a parametric ANOVA analysis in a multi-group comparison using the XCMS server software tool. In total, we were able to align, on average 268 m/z peak-retention time pairs in each sample, reflecting the complex chemical landscape within the cultures. To reduce this complexity, principal component analysis was performed that summarized 55% of the culture variability in the GC/MS data by the first two principal components (PC): PC1, 39%; PC2, 16%. The principal component analysis separated the four exchange experiments into two defined groups along axis 2 (Figure 3a). While the Acidithrix cultures [Acx (Acx); Acx (Acp)] had a positive PC2, the Acidiphilium cultures [Acp (Acx); Acp (Acp)] resulted in a negative PC2. In summary, principal component analysis highlights strong differences in the chemical composition between cultures of each bacterial strain. Incubation with cell-free supernatant of the respective partner did not result in characteristic changes of the metabolome of the respective species.
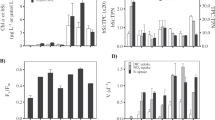
Statistical analysis, structure elucidation and quantification of components from spent bacterial medium. (a) Principal component analysis scores comparing the four types of bacterial cultures using a multi-group comparison with XCMS. The size of the spots represents the unexplained variation. Two groups reflecting the respective bacterial species are labeled with ellipsoids. (b) Cloud-plot analysis of the multi-group comparison of relevant features from XCMS analysis of the four types of bacterial cultures. Each cloud represents a feature that differs between the cultures, plotted by the retention time (x-axis) and the mass to charge (m/z) ratio in the mass spectrum (y-axis). The color indicates the features that were prevalent in Acidithrix strain C25 cultures (red) or in Acidiphilium strain C61 cultures (blue, corresponding to the grouping in a). The size of the clouds indicates the fold change of the features; the color depth the statistical relevance. (c) Assigned compounds to the hits found by XCMS using the NIST GC/MS library. All compounds were identified as bis-trimethylsilyl derivatives and here the native compounds are shown. (d) Quantity of each compound in the four types of bacterial cultures. The absolute values of phenethylamine (d1) were measured with an additional ultra high performance liquid chromatography/high resolution mass spectrometry method. p-Hydroxybenzoic acid (d2) and furane 2,5-dicarboxylic acid (d3) were relatively quantified from GC/MS measurements.
Further analysis was conducted using a cloud-plot analysis, which is used to indicate the mass-retention time pairs, called features that differ between the compared sample sets. Figure 3b shows most significant metabolites specific for the respective species. Due to fragmentation of ions in EI ionization GC/MS, cloud-plot analyses give several clouds at different m/z values at one single retention time that can be summarized in a feature group corresponding to one metabolite. Within the monitored retention time window (10 and 48 min), we were able to detect, in total, 10 relevant feature groups. Six of these features were specifically prevalent in Acidithrix cultures (red color, Figure 3b) and four in Acidiphilium cultures (blue color, Figure 3b). The features detected within the GC/MS data were further analyzed with the NIST EI-MS library. We were able to identify 4 compounds out of the 10 relevant feature groups: 2-phenethyamine (PEA), p-hydroxybenzoic acid, furane 2,5-dicarboxylic acid and the amino acid tryptophan (Figure 3c). To verify the chemical identity of these hits, we subjected standards of the four compounds to derivatization and GC/MS analysis. Standards and analytes had the same retention time and gave identical mass spectra, thereby unambiguously proving the structural identity. The other remaining six feature groups did not match any result within the NIST library. For quantification of PEA, solid-phase extraction proved not to be suitable, since the primary amine shows variable extraction success in the acidic culture broth. To overcome this limitation we utilized a short ultra high performance liquid chromatography/high resolution mass spectrometry analysis of the supernatant from the (co-)cultures yielding concentrations of PEA in Acx (Acx) and Acx (Acp) of 10.5±5.4 μM and 2.1±5.5 μM, respectively. The concentration was below the detection limit in the other incubations (Figure 3d1). p-Hydroxybenzoic acid was relatively more abundant in the cultures of Acidithrix and furane 2,5-dicarboxylic acid was more abundant in the cultures of Acidiphilium (relative quantification in solid-phase extraction extract, Figures 3d2 and d3).
Screening of possible allelochemicals
To elucidate the potential ecological role of the identified species-specific metabolites, commercial grade pure PEA, p-hydroxybenzoic acid and furane 2,5-dicarboxylic acid were added to Acidithrix strain C25 and Acidiphilium strain C61 cultures at concentrations of 1–1000 μM. Aggregation similar to that in Acp (Acx) incubations was observed in pure cultures of Acidiphilium supplemented with 5 μM or higher PEA concentrations following 1-week incubation (Supplementary Figure S2). The active concentration is thus even below the PEA content of Acx (Acx) incubations (Figure 3d). Quantitative PCR, a more sophisticated method for determining differences in bacterial abundance compared with total nucleic acid measurements, revealed significantly higher bacterial 16S rRNA gene copy numbers in pure culture of Acidiphilium following the addition of 10 μM PEA and slightly lower in cultures supplemented with 100 μM PEA when compared with the control incubations without exogenous PEA following a 6-day incubation period (Figure 4). No effect on cell morphology of Acidiphilium was observed following the addition of either p-hydroxybenzoic acid or furane 2,5-dicarboxylic acid, and none of these chemicals affected growth of Acidithrix.
Bacterial 16S rRNA gene copy numbers in Acidiphilium strain C61 cultures with or without addition of exogenous PEA. Copy numbers ml−1 of bacterial 16S rRNA gene on day 6 of incubation were quantified and compared among Acidiphilium strain C61 cultures without supply of PEA, with 10 μM PEA and 100 μM PEA. n=9, *P<0.05, **P<0.01 (Welch’s t-test), error bars indicate standard deviations.
Direct physical interactions between Acidithrix and Acidiphilium
To determine direct interaction between both strains, Acidiphilium was inoculated to a pre-grown culture of Acidithrix. After 3–4 days of co-cultivation, putative cell aggregates of Acidiphilium were formed and appeared to co-exist with filamentous Acidithrix cells (Figure 5). These aggregates showed the same morphology as those triggered by cell-free Acidithrix supernatant or pure PEA.
Microscopic images of co-culture incubations of Acidithrix strain C25 and Acidiphilium strain C61. (Co-)cultures were visualized with nucleic acid staining (SYTO 13). The figures show pure cultures of Acidithrix strain C25 (left) and Acidiphilium strain C61 (right). Co-cultures (middle) show characteristic aggregates of Acidiphilium that are also observed in treatments with PEA.
Discussion
Previous studies involving the interactions of acidophilic microorganisms focused on gene regulation as the foundations for interactions or consumption of inhibitory compounds (Liu et al., 2011). Our study highlights the functional basis for microbial interactions by directly targeting the metabolites mediating these exchanges within iron snow aggregates. These chemical mediators not only increase the growth of Acidithrix and Acidiphilium in exchange experiments, but might also affect the rates of Fe-cycling in iron snow.
We could show that the rate of Fe(II) oxidation by Acidithrix strain C25 was enhanced when grown in the presence of Acidiphilium strain C61 cell-free supernatant. In addition, we observed increases in the amounts of total nucleic acid in Acidithrix strain C25 cultures grown in presence of Acidiphilium strain C61 cell-free supernatant, suggesting that chemicals produced and secreted by Acidiphilium stain C61 positively affected activity and biomass production of Acidithrix strain C25. Similarly, the chemolithoautotrophic Acidithiobacillus ferrooxidans shows increased rates of Fe(II) oxidation and greater cell densities when co-cultured with the heterotroph Acidiphilium acidophilum (Liu et al., 2011). These effects were attributed to the activation of Fe(II) oxidation-related genes and a set of RuBisCO-encoding genes likely due to consumption of substrates by the heterotroph A. acidophilum that typically function to inhibit Fe(II) oxidation (Liu et al., 2011). However, this mechanism is not applicable in our supernatant exchange experiment. More likely, chemical mediators produced by the heterotroph Acidiphilium strain C61 and present in the cell-free supernatant stimulated growth of Acidithrix strain C25.
PEA as the aggregation signal
In most Acidiphilium species, cell motility is controlled by polar flagella (Harrison, 1981; Wichlacz et al., 1986). The flagellar-mediated cell motility typically prevents cell aggregation (Caldara et al., 2012). However, Acidiphilium stain C61 cells formed distinct macroscopic aggregates following addition of either Acidithrix strain C25, cell-free supernatant of this strain or pure PEA in concentrations comparable to those in cultures. This clearly demonstrates that PEA acts as a chemical mediator triggering cell aggregation by Acidiphilium stain C61 and thus the formation of iron snow.
In another gram-negative bacterium, Proteus mirabilis, PEA is involved in the cell transition cycle (Stevenson and Rather, 2006). PEA signals P. mirabilis cells to shift from swarming to vegetative state by inhibiting the assembly or activity of FlhDC, a key regulator of flagellin expression of lateral (peritrichous) flagellum systems and swarmer cell differentiation (Stevenson and Rather, 2006). However, swarming activity of P. mirabilis cultures is inhibited by 50% at 1 mM PEA and complete inhibition was observed under the influence of 4 mM PEA. Inhibition of swarming in P. mirabilis induces cell transition into the vegetative phase, therefore PEA functions as a consolidation signal for the cells that lead to accelerated cell multiplication (Matsuyama et al., 2000). In our study, cell aggregation of Acidiphilium was induced following the addition of 5–10 μM of exogenous PEA, which were in the concentration range of PEA detected in the supernatant of Acidithrix cultures. Our study also revealed that PEA induces faster growth of Acidiphilium cells at low in situ relevant concentrations of 10 μM.
The observed aggregation of Acidiphilium could be triggered by inhibition of flagellar-mediated cell motility by the presence of PEA. According to the genome database on Integrated Microbial Genomes (Markowitz et al., 2012), all Acidiphilium genera within the database lack the flhDC gene cluster, indicating the absence of a peritrichous flagellum system in Acidiphilium. The type strain of Acidiphilium cryptum was reported to possess one polar flagellum or two lateral flagella (Harrison, 1981), which are supposed to be regulated by the sigma factor 54-dependent NtrC family of transcriptional activators for flagellum gene expression (Arora et al., 1997; Jyot et al., 2002; Soutourina and Bertin, 2003). Differences in the flagellum regulatory systems between Acidiphilium strain C61 and P. mirabilis might explain the very low concentration of PEA necessary to inhibit Acidiphilium cell motility.
Metabolomics analyses in our study detected the presence of PEA in Acidithrix cultures but not in Acidiphilium cultures even if they were supplemented with PEA containing supernatant of Acidithrix. In addition, PEA was not detected in cell-free controls amended with supernatant of Acidithrix after 10 days incubation. This suggests that PEA produced by Acidithrix and derived from their supernatant did not remain over the 10-day incubation period due to either abiotic degradation or consumption by Acidiphilium. However, this result also indicates that PEA was produced by Acidithrix cells over the entire bioassay. Microscopic images of Acidithrix and Acidiphilium grown in co-culture (Figure 5) show that cell aggregation of Acidiphilium was induced in the presence of Acidithrix. The similarity of aggregate morphology in co-cultures and PEA-treated Acidiphilium, as well as the concentrations of PEA observed in cultures, supports this aromatic amine to be the exclusive active factor. Interestingly, in presence of Acidiphilium cell-free supernatant, PEA production by Acidithrix was lower than the PEA production observed in Acx (Acx) incubations. This result implies the presence of a two-way signal transduction system may potentially function to mediate chemical signaling when Acidithrix and Acidiphilium grow in close proximity to one another, thereby regulating the production, detection and utilization of PEA.
Previous studies have reported PEA production by algae, plants, fungi and bacteria (Irsfeld et al., 2013). The biosynthesis of PEA typically occurs via decarboxylation of L-phenylalanine by the aromatic L-amino acid decarboxylase (Irsfeld et al., 2013). However, L-phenylalanine decarboxylation pathways and mechanisms in bacteria have not been well characterized, and only small groups of food fermentation-related gram-positive bacteria were reported to produce PEA by enzymatic activities also involving the decarboxylation of tyrosine to tyramine (Pessione et al., 2009; Marcobal et al., 2012). According to the Integrated Microbial Genomes database, the type strain Acidithrix ferrooxidans Py-F3 lacks any homologous gene sequence encoding the tyrosine/phenylalanine decarboxylase genes found in either the genome of P. mirabilis or the genomes of food fermenting bacteria, possibly due to the incomplete genome sequence. The biosynthetic pathway responsible for PEA production in Acidithrix strain C25 has not been elucidated, so the exact mechanisms regulating the chemical signaling pathways in these two partner bacteria require further investigations.
Model for the chemical communication between Acidithrix and Acidiphilium in iron snow
Acidithrix strain C25 was suggested to be involved in early stages of iron snow formation via Fe(II) oxidation and subsequent aggregate formation associated with iron minerals (Mori et al., 2016), followed by colonization of Acidiphilium and other heterotrophs, chemolithoautotrophs and photoautotrophs on the aggregates.
Apparently, motile Acidiphilium cells rapidly colonize microbe-mineral assemblages in the iron snow to obtain organic carbon sources as well as oxidized Fe, which can be used by the bacteria as an electron acceptor both under oxic and anoxic conditions (Küsel et al., 2002). According to our results, these microorganisms lose their motility, attach and ultimately form aggregates on the surface of iron snow particles under the influence of PEA produced by Acidithrix. PEA additionally acts as the consolidation signal that induces faster cell growth by the Acidiphilium cells associated with the iron snow aggregates (Figure 6). Colonization of Acidiphilium benefits Acidithrix by inducing enhanced growth and Fe(II) oxidation rates.
Putative schematic model depicting colonization of Acidithrix strain C25 and Acidiphilium strain C61 within iron snow particles induced by phenethylamine (PEA). (a) Filamentous Acidithrix strain C25 cells form aggregates in association with solid Fe(III) particles and PEA accumulates around these aggregates. (b) Motility of free-living Acidiphilium strain C61 cells is inhibited under the influence of PEA, which induces faster multiplication and trigger cell colonization. (c) A larger cell-mineral aggregate is formed and precipitates to deeper water layers.
In summary, we show that PEA functions as the allelochemical between Acidithrix and Acidiphilium, which promotes rapid co-colonization onto the surface of the iron snow particles. Undoubtedly, the short residence time of iron snow in the water column of these shallow lakes (Reiche et al., 2011) does not allow all of the potentially complex aspects of biological activities and trophic interactions to occur, which is reflected by the community structure of the iron snow dominated by some groups of Fe(II) oxidizers and Fe(III) reducers (Lu et al., 2013). The interspecies cell signaling between the model isolates of this study may have evolved to allow both bacterial strains to capitalize on the strengths of the partner and to access the polycrystalline schwertmannite (Miot et al., 2016), the main Fe mineral of the iron snow aggregates. The interaction we observed can be attributed to the theory of microbial chemical communication, such that both the emitter and the receiver gain benefits through production, secretion and utilization of chemical signals mediating a beneficial partnership that can remain stable over evolutionary time (Keller and Surette, 2006). The stability of the dominant bacterial community associated with iron snow particles (Lu et al., 2013) suggests the observed interactions between these bacteria have evolved specifically such that the Fe(II) oxidizer and Fe(III) reducer can thrive in acidic lakes. Future studies are expected to reveal the biosynthetic pathway and functional mechanisms of PEA, as well as its effect on other dominant bacterial species that comprise the iron snow surface-associated microbial community, such as Ferrovum sp. (Lu et al., 2013). These studies will provide further insight into interspecies cell–cell chemical communication in acidic, Fe-rich aquatic environments.
References
Alldredge AL, Silver MW . (1988). Characteristics, dynamics and significance of marine snow. Prog Oceanogr 20: 41–82.
Arora SK, Ritchings BW, Almira EC, Lory S, Ramphal R . (1997). A transcriptional activator, FleQ, regulates mucin adhesion and flagellar gene expression in Pseudomonas aeruginosa in a cascade manner. J Bacteriol 179: 5574–5581.
Caldara M, Friedlander Ronn S, Kavanaugh Nicole L, Aizenberg J, Foster Kevin R, Ribbeck K . (2012). Mucin biopolymers prevent bacterial aggregation by retaining cells in the free-swimming state. Curr Biol 22: 2325–2330.
Dang H, Lovell CR . (2016). Microbial surface colonization and biofilm development in marine environments. Microbiol Mol Biol Rev 80: 91–138.
Fenchel T . (2001). Eppur si muove: many water column bacteria are motile. Aquat Microb Ecol 24: 197–201.
Giani M, Rinaldi A, Degobbis D . (2005). Mucilages in the Adriatic and Tyrrhenian Sea: an introduction. Sci Total Environ 353: 3–9.
Gillard J, Frenkel J, Devos V, Sabbe K, Paul C, Rempt M et al. (2013). Metabolomics enables the structure elucidation of a diatom sex pheromone. Angew Chem Int Ed 52: 854–857.
Gram L, Grossart HP, Schlingloff A, Kiørboe T . (2002). Possible quorum sensing in marine snow bacteria: production of acylated homoserine lactones by Roseobacter strains isolated from marine snow. Appl Environ Microbiol 68: 4111–4116.
Grossart HP, Simon M . (1998). Bacterial colonization and microbial decomposition of limnetic organic aggregates (lake snow). Aquat Microb Ecol 15: 127–140.
Harrison AP . (1981). Acidiphilium cryptum gen. nov., nov., heterotrophic bacterium from acidic mineral environments. Int J Syst Bacteriol 31: 327–332.
Irsfeld M, Spadafore M, Prüß BM . (2013). β-phenylethylamine, a small molecule with a large impact. WebmedCentral 4: 4409.
Jatt AN, Tang K, Liu J, Zhang Z, Zhang XH . (2015). Quorum sensing in marine snow and its possible influence on production of extracellular hydrolytic enzymes in marine snow bacterium Pantoea ananatis B9. FEMS Microbiol Ecol 91: 1–13.
Johnson DB, Hallberg KB . (2007) Techniques for detecting and identifying acidophilic mineral-oxidizing microorganisms. In: Rawlings DE, Johnson DB (eds). Biomining. Springer: Berlin, Germany, pp 237–261.
Johnson M, Zaretskaya I, Raytselis Y, Merezhuk Y, McGinnis S, Madden TL . (2008). NCBI BLAST: a better web interface. Nucleic Acids Res 36: W5–W9.
Jones RM, Johnson DB . (2015). Acidithrix ferrooxidans gen. nov., sp. nov.;, a filamentous and obligately heterotrophic, acidophilic member of the Actinobacteria that catalyzes dissimilatory oxido-reduction of iron. Res Microbiol 166: 111–120.
Jyot J, Dasgupta N, Ramphal R . (2002). FleQ, the major flagellar gene regulator in Pseudomonas aeruginosa, binds to enhancer sites located either upstream or atypically downstream of the RpoN binding site. J Bacteriol 184: 5251–5260.
Keller L, Surette MG . (2006). Communication in bacteria: an ecological and evolutionary perspective. Nat Rev Microbiol 4: 249–258.
Kiørboe T, Tang K, Grossart H-P, Ploug H . (2003). Dynamics of microbial communities on marine snow aggregates: colonization, growth, detachment, and grazing mortality of attached bacteria. Appl Environ Microbiol 69: 3036–3047.
Kuhlisch C, Pohnert G . (2015). Metabolomics in chemical ecology. Nat Prod Rep 32: 937–955.
Küsel K, Dorsch T, Acker G, Stackebrandt E . (1999). Microbial reduction of Fe(III) in acidic sediments: isolation of Acidiphilium cryptum JF-5 capable of coupling the reduction of Fe(III) to the oxidation of glucose. Appl Environ Microbiol 65: 3633–3640.
Küsel K, Roth U, Drake HL . (2002). Microbial reduction of Fe(III) in the presence of oxygen under low pH conditions. Environ Microbiol 4: 414–421.
Lane DJ . (1991). 16S/23S rRNA sequencing. In Stackebrandt E, Goodfellow M eds Nucleic Acid Techniques in Bacterial Systematics Wiley: New York, NY, USA, pp 115–175.
Liu H, Yin H, Dai Y, Dai Z, Liu Y, Li Q et al. (2011). The co-culture of Acidithiobacillus ferrooxidans and Acidiphilium acidophilum enhances the growth, iron oxidation, and CO2 fixation. Arch Microbiol 193: 857–866.
Long RA, Azam F . (2001). Antagonistic interactions among marine pelagic bacteria. Appl Environ Microbiol 67: 4975–4983.
Lu S, Chourey K, Reiche M, Nietzsche S, Shah MB, Neu TR et al. (2013). Insights into the structure and metabolic function of microbes that shape pelagic iron-rich aggregates (‘iron snow’). Appl Environ Microbiol 79: 4272–4281.
Marcobal A, De Las Rivas B, Landete JM, Tabera L, Muñoz R . (2012). Tyramine and phenylethylamine biosynthesis by food bacteria. Crit Rev Food Sci Nutr 52: 448–467.
Markowitz VM, Chen I-MA, Palaniappan K, Chu K, Szeto E, Grechkin Y et al. (2012). IMG: the integrated microbial genomes database and comparative analysis system. Nucleic Acids Res 40: D115–D122.
Matsuyama T, Takagi Y, Nakagawa Y, Itoh H, Wakita J, Matsushita M . (2000). Dynamic aspects of the structured cell population in a swarming colony of Proteus mirabilis. J Bacteriol 182: 385–393.
Miot J, Lu S, Morin G, Adra A, Benzerara K, Küsel K . (2016). Iron mineralogy across the oxycline of a lignite mine lake. Chem Geol 434: 28–42.
Mori JF, Lu S, Händel M, Totsche KU, Neu TR, Iancu VV et al. (2016). Schwertmannite formation at cell junctions by a new filament-forming Fe(II)-oxidizing isolate affiliated with the novel genus Acidithrix. Microbiology 162: 62–71.
Passow U, Ziervogel K, Asper V, Diercks A . (2012). Marine snow formation in the aftermath of the Deepwater Horizon oil spill in the Gulf of Mexico. Environ Res Lett 7: 035301.
Paul C, Mausz MA, Pohnert G . (2013). A co-culturing/metabolomics approach to investigate chemically mediated interactions of planktonic organisms reveals influence of bacteria on diatom metabolism. Metabolomics 9: 349–359.
Pessione E, Pessione A, Lamberti C, Coïsson DJ, Riedel K, Mazzoli R et al. (2009). First evidence of a membrane-bound, tyramine and β-phenylethylamine producing, tyrosine decarboxylase in Enterococcus faecalis: a two-dimensional electrophoresis proteomic study. Proteomics 9: 2695–2710.
Prince EK, Pohnert G . (2010). Searching for signals in the noise: metabolomics in chemical ecology. Anal Bioanal Chem 396: 193–197.
Reiche M, Lu S, Ciobota V, Neu TR, Nietzsche S, Rösch P et al. (2011). Pelagic boundary conditions affect the biological formation of iron-rich particles (iron snow) and their microbial communities. Limnol Oceanogr 56: 1386–1398.
Simon M, Grossart HP, Schweitzer B, Ploug H . (2002). Microbial ecology of organic aggregates in aquatic ecosystems. Aquat Microb Ecol 28: 175–211.
Soutourina OA, Bertin PN . (2003). Regulation cascade of flagellar expression in gram-negative bacteria. Fems Microbiol Rev 27: 505–523.
Stevenson LG, Rather PN . (2006). A novel gene involved in regulating the flagellar gene cascade in Proteus mirabilis. J Bacteriol 188: 7830–7839.
Suess E . (1980). Particulate organic carbon flux in the oceans—surface productivity and oxygen utilization. Nature 288: 260–263.
Tamura H, Goto K, Yotsuyanagi T, Nagayama M . (1974). Spectrophotometric determination of iron(II) with 1,10-phenanthroline in presence of large amounts of iron(III). Talanta 21: 314–318.
Tautenhahn R, Patti GJ, Rinehart D, Siuzdak G . (2012). XCMS online: a web-based platform to process untargeted metabolomic data. Anal Chem 84: 5035–5039.
Thornton D . (2002). Diatom aggregation in the sea: mechanisms and ecological implications. Eur J Phycol 37: 149–161.
Tischler JS, Jwair RJ, Gelhaar N, Drechsel A, Skirl A-M, Wiacek C et al. (2013). New cultivation medium for ‘Ferrovum’ and Gallionella-related strains. J Microbiol Methods 95: 138–144.
Wichlacz PL, Unz RF, Langworthy TA . (1986). Acidiphilium angustum sp. nov., Acidiphilium facilis sp. nov., and Acidiphilium rubrum sp. nov.: acidophilic heterotrophic bacteria isolated from acidic coal mine drainage. Int J Syst Evol Microbiol 36: 197–201.
Zan J, Liu Y, Fuqua C, Hill RT . (2014). Acyl-homoserine lactone quorum sensing in the Roseobacter clade. Int J Mol Sci 15: 654–669.
Acknowledgements
Support for this research was kindly provided by the Deutsche Forschungsgemeinschaft (DFG) to the German Centre for Integrative Biodiversity Research (iDiv) Halle-Jena-Leipzig, the research training group GRK 1257, which is part of the Jena School for Microbial Communication (JSMC), the collaborative research center 1076 AquaDiva, and the collaborative research center 1127 Chemical Mediators in Complex Biosystems (ChemBioSys). We are grateful to Prof Dr Susan E Trumbore for helpful advice and comments on the manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on The ISME Journal website .
Supplementary information
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/4.0/
About this article
Cite this article
Mori, J., Ueberschaar, N., Lu, S. et al. Sticking together: inter-species aggregation of bacteria isolated from iron snow is controlled by chemical signaling. ISME J 11, 1075–1086 (2017). https://doi.org/10.1038/ismej.2016.186
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ismej.2016.186
This article is cited by
-
The non-attached biofilm aggregate
Communications Biology (2023)
-
Estuarine gradients dictate spatiotemporal variations of microbiome networks in the Chesapeake Bay
Environmental Microbiome (2021)
-
Burial of microplastics in freshwater sediments facilitated by iron-organo flocs
Scientific Reports (2021)
-
Iron is not everything: unexpected complex metabolic responses between iron-cycling microorganisms
The ISME Journal (2020)
-
Potential role of N-acetyl glucosamine in Aspergillus fumigatus-assisted Chlorella pyrenoidosa harvesting
Biotechnology for Biofuels (2019)