Abstract
Bacterial type VI secretion systems (T6SSs) are molecular weapons designed to deliver toxic effectors into prey cells. These nanomachines have an important role in inter-bacterial competition and provide advantages to T6SS active strains in polymicrobial environments. Here we analyze the genome of the biocontrol agent Pseudomonas putida KT2440 and identify three T6SS gene clusters (K1-, K2- and K3-T6SS). Besides, 10 T6SS effector–immunity pairs were found, including putative nucleases and pore-forming colicins. We show that the K1-T6SS is a potent antibacterial device, which secretes a toxic Rhs-type effector Tke2. Remarkably, P. putida eradicates a broad range of bacteria in a K1-T6SS-dependent manner, including resilient phytopathogens, which demonstrates that the T6SS is instrumental to empower P. putida to fight against competitors. Furthermore, we observed a drastically reduced necrosis on the leaves of Nicotiana benthamiana during co-infection with P. putida and Xanthomonas campestris. Such protection is dependent on the activity of the P. putida T6SS. Many routes have been explored to develop biocontrol agents capable of manipulating the microbial composition of the rhizosphere and phyllosphere. Here we unveil a novel mechanism for plant biocontrol, which needs to be considered for the selection of plant wardens whose mission is to prevent phytopathogen infections.
Similar content being viewed by others
Introduction
The type VI secretion system (T6SS) is found in more than 25% Gram-negative bacteria and used to inject toxic proteins into prokaryotic or eukaryotic cells (Ho et al., 2013). Initially, the T6SS was assigned a role in virulence and eukaryotic cell manipulation (Ma and Mekalanos, 2010; Miyata et al., 2011). Further analyses showed that this system has a key role in inter-bacterial competition (Ho et al., 2013). It provides selective advantages to producer strains by annihilating competitors either in an indiscriminate manner or in response to danger signals (Hood et al., 2010; Basler et al., 2013; Ho et al., 2013; Hachani et al., 2014). The T6SS toxins are usually produced together with immunity proteins that prevent self-intoxication. In molecular terms, the T6SS displays structural similarities with the tail and the puncturing device of the bacteriophage T4 (Leiman et al., 2009; Filloux, 2011; Cascales and Cambillau, 2012). It is composed by 13 core components of which some have now been assigned clear functions (Figure 1a). TssB and TssC form a contractile sheath that encases a tube formed by rings of Hcp hexamers (Kudryashev et al., 2015). A puncturing device made up of a trimeric VgrG spike is placed on top of the Hcp tube and crowned with a PAAR protein (Cascales and Cambillau, 2012; Shneider et al., 2013). The cytosolic part of the T6SS docks onto a membrane complex (TssLMJ) probably by interacting with a phage baseplate-like structure (Durand et al., 2015; Filloux and Freemont, 2016; Planamente et al., 2016). On contraction of the TssBC sheath, the T6SS effectors are propelled out of the bacterium together with the Hcp and VgrG proteins and delivered into prey cells (Basler and Mekalanos, 2012). Finally, the ClpV ATPase binds the contracted sheath for disassembly and recycling, enabling another round of injection (Kapitein et al., 2013; Kube et al., 2014). The T6SS is usually quite modular and can accommodate different combinations of VgrG/PAAR proteins to form the tip. This modularity allows the delivery of a great variety of effectors (Shneider et al., 2013; Hachani et al., 2014; Whitney et al., 2014). Alternatively, the effectors can also be ushered in and delivered by the tube-forming Hcp protein (Silverman et al., 2013). Thus, a wide variety of effectors with a broad range of activities can be secreted by a single T6SS. T6SS effectors have been classified into specialized and cargo effectors (Cianfanelli et al., 2016). Specialized effectors are domains, usually at the C-terminus of specific T6SS structural components that are coined as ‘evolved’ VgrG, PAAR or Hcp proteins. In contrast, cargo effectors interact non-covalently with ‘canonical’ VgrG, PAAR or Hcp proteins (Durand et al., 2014). Several cargo effectors carry a motif named MIX (marker for type six effectors) that is proposed to be required for recognition and direct interaction with VgrG or PAAR proteins (Salomon et al., 2014). Specific adaptors such as Tap/Tec and EagR proteins are likely to facilitate the interaction between the structural components of the T6SS tip and the effectors (Alcoforado Diniz and Coulthurst, 2015; Liang et al., 2015; Unterweger et al., 2015). Finally, accessory elements (named tag from T6SS accessory genes) are required to modulate the assembly of the system and/or confer additional functions (Boyer et al., 2009).
T6SS clusters in P. putida KT2440. (a) Schematic representation of the T6SS structure. (b) Distribution of the K1-, K2- and K3-T6SS clusters (blue), and the vgrG (yellow) and hcp (purple) genes in the KT2440 genome. (c–e) Genomic organization of the P. putida T6SSs cluster, including K1 (c), K2 and K3 (d) or the vgrG and hcp orphan clusters (e). The color code of the genes correlates with the color code shown in a. The asterisk (*) in the tssC2, vgrG2 and hcp4 genes indicates that these genes contain premature stop codons.
The T6SS was first identified in two pathogenic bacteria, Vibrio cholerae and Pseudomonas aeruginosa (Mougous et al., 2006; Pukatzki et al., 2006) and analyzed later in many other pathogens (Suarez et al., 2008; de Pace et al., 2010; Burtnick et al., 2011; Murdoch et al., 2011; Rosales-Reyes et al., 2012; Lin et al., 2013). However, analytical description of T6SS in non-pathogenic bacteria is underrepresented in the literature (Bladergroen et al., 2003; Marchi et al., 2013), despite an even distribution in both classes of organisms (Boyer et al., 2009). Pseudomonas putida is a saprophytic soil bacterium that has the capacity to colonize the root of crop plants (Espinosa-Urgel et al., 2000; Molina et al., 2000). It is a well-established biocontrol agent that provides growth advantages to the plant (Weller, 2007). In this study we identified and characterized the P. putida T6SS, which exhibits great variety and complexity both in terms of apparatus and secreted toxins. We showed that this secretion system is used by the bacterium to drive killing of resilient phytopathogens and appears to be a major factor in its biocontrol portfolio.
Materials and methods
Bacterial strains and growth conditions
Bacterial strains are listed in Supplementary Table S1. P. putida mutants were constructed by allelic exchange as described previously (Vasseur et al., 2005). Briefly, 750 bp DNA fragments upstream and downstream the gene to be deleted were amplified using KT2440 genomic DNA. Mutator fragments were obtained by overlapping PCR, cloned into pCR-BluntII-TOPO (Invitrogen, ThermoFisher Scientific, Loughborough, UK), sequenced and subcloned into the pKNG101 suicide vector (Kaniga et al., 1991). A similar approach was used to replace the wild-type tke2 gene with tke2-V5 encoding a C-terminally virus 5 (V5)-tagged Tke2 protein. The hcp1-HA gene encoding a C-terminal hemagglutinin (HA)-tagged Hcp1 protein was introduced on the chromosome using the miniCTX transposon (Hoang et al., 2000). Insertions and gene replacements were confirmed by PCR. All strains were grown in lysogeny broth (LB; Sambrook et al., 1989). For secretion assays, tryptone soya broth medium (Oxoid, ThermoFisher Scientific, Loughborough, UK) was used. Escherichia coli was incubated at 37 °C, and P. putida and the phytopathogens at 25–30 °C. Antibiotics were used at (μg ml−1): ampicillin, 100; gentamycin, 20; kanamycin, 50; piperacillin, 25; rifampicin, 20; streptomycin, 100; tetracycline, 50.
Plasmids and cloning
Plasmids are described in Supplementary Table S1 and primers listed in Supplementary Table S2. PCR amplifications were performed using Phusion Hot Start High-Fidelity (Finnzymes, ThermoFisher Scientific, Loughborough, UK), KOD Hot Start (EMD Millipore, Watford, UK) or Taq (Roche, Burgess Hill, UK) DNA polymerases. Recombinant plasmids were sequenced and transferred to P. putida by electroporation (Choi et al., 2006) or conjugation (Ramos-Gonzalez et al., 1991).
Bioinformatic analyses
Pseudomonas sequences were obtained from the Pseudomonas Genome database (Winsor et al., 2016). BLASTP analyses were performed at the NCBI website (Boratyn et al., 2013) and amino acid sequence searches using SMART (Letunic et al., 2015) and Pfam (Finn et al., 2016). The Protein Homology/analogy Recognition Engine (Phyre2) server was used to perform structural-base homology prediction (Kelley et al., 2015). The PyMOL Molecular Graphics System (Version 1.8 Schrondinger, LLC, Cambridge, UK) was used to build structural alignments. The phylogenetic tree was constructed using MEGA6 (Tamura et al., 2013). PSORTb software and SOSUI GramN server were used to predict sub-cellular location of proteins (Imai et al., 2008; Yu et al., 2010), TMHMM software to predict transmembrane domains (Krogh et al., 2001), and SignalP and SOSUIsignal to predict signal peptides (Gomi et al., 2004; Petersen et al., 2011). Synteny was analyzed using the CoGe’s Genome Evolution tool (Lyons and Freeling, 2008). The UGENE bioinformatics software was used to identify open reading frames (orfs; Okonechnikov et al., 2012).
Secretion assays
Bacterial strains were grown in tryptone soya broth for 5 h at 30 °C and the extracellular fraction obtained and analyzed as previously described (Hachani et al., 2011). The proteins in the culture supernatants were precipitated with trichloroacetic acid and resuspended in 1 M of Tris-base and 4 × Laemmli buffer. Proteins were separated by SDS–polyacrylamide gel electrophoresis containing 8% or 15% (w/v) acrylamide and electro-transferred to nitrocellulose membranes. Immunodetection was performed using monoclonal antibodies directed against the influenza HA epitope (HA.11, Covance, Biolegend, London, UK) or the paramyxovirus of simian V5 epitope (Invitrogen, ThermoFisher Scientific). A monoclonal antibody against the β-subunit of the RNA polymerase (Neoclone, Biolegend, London, UK) was also used. The secondary antibody, horseradish peroxidase-conjugated rabbit anti-mouse (Sigma Aldrich, Gillingham, UK), was detected using the SuperSignal West Pico Chemiluminescent Substrate (Thermo Scientific, Loughborough, UK). Blots were scanned and analyzed using the Image Reader LAS-3000 (Fuji, GE Healthcare, Little Chalfont, UK).
Growth inhibition assays
Overnight LB cultures of E. coli DH5α harboring the vectors pNDM220 (Gotfredsen and Gerdes, 1998), pBAD33 (Guzman et al., 1995) or derivatives encoding Tke2 or Tki2 were adjusted to OD600 of 0.1. Expression of tke2 and tki2 was induced with 0.2% (w/v) L-arabinose and 1 mM isopropyl β-D-1-thiogalactopyranoside, respectively.
Interbacterial competition assays
In vitro competition assays were performed on LB plates as previously described (Hachani et al., 2013). Bacterial overnight cultures were adjusted to OD600 of 1 in PBS and mixed in a 1:1 ratio (P. putida-prey). Bacteria were co-cultured at 30 °C for 5 h (E. coli) or 24 h (phytopathogens). The competition was quantified by counting colony-forming units on antibiotic selection. At least three biologically independent experiments were performed. In planta competition assays were carried out by infiltration of bacteria into Nicotiana benthamiana leaves as described before (Ma et al., 2014). Overnight cultures of P. putida and Xanthomonas campestris were adjusted to OD600 of 0.1 in PBS and mixed in a 1:1 ratio. Approximately 100 μl volume was infiltrated on the reverse of a 1-month-old leaf and the infiltration area marked. After 24 h of incubation in a plant chamber (23 °C, 16 h light), colony-forming units were determined. A section of the leaf from the infiltration area was cut out, homogenized in PBS and subsequently serially diluted. The leaves were visualized by fluorescence microscopy using a Leica M206FA stereomicroscope. Imaging was performed at room temperature with a × 1 objective. The evaluation of necrosis was based on the coloration of the leaves following previous standard evaluation of virulence that goes from no visible effects to changes in the tissue color of the leaf, which can shift from green to yellowish (chlorosis), yellowish to brownish and blackening of the leaf (necrosis), up to complete rotting of the leaf at later stages (Katzen et al., 1998). In our assay, the circled zones point at deep brown color area.
Results
Genome-wide screening for T6SSs in P. putida species
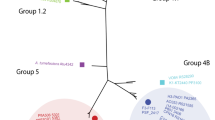
In silico analyses of forty P. putida strains revealed that all encode T6SS genes and 90% of them have at least one cluster encoding a full set of T6SS components (Supplementary Table S3). The number of T6SS clusters in a single strain ranged from zero in P. putida BIRD-1 or H8234 to four in P. putida PA14H7, whereas most strains contained one or two clusters (Supplementary Table S3). In total, we identified 66 complete T6SS gene clusters distributed in three main phylogenetic clades (Figure 2). Following previous nomenclature (Boyer et al., 2009; Barret et al., 2011), we referred to these three groups as 1.2, 2 and 4B. Eighty percent of the clusters belong to group 1.2 or 4B, whereas 10% are found in group 2 (Figure 2). Each of these groups contains distinguishable genetic architecture and features (Supplementary Figure S1), as described in the next section.
Phylogenetic distribution of T6SS clusters in P. putida species. Maximum likelihood tree with 1000 bootstrap replicates were built with Mega 6 for the core component protein TssB. T6SS cluster nomenclature (Boyer et al., 2009; Barret et al., 2011) is used to show the major phylogenetic clusters. Three main groups are clearly distinguishable: group 1.2 (green), group 2 (red) and group 4B (blue). P. aeruginosa and A. tumefaciens T6SSs loci are included into the phylogenetic tree to illustrate all the subgroups: 1.1 (P. aeruginosa H2), 1.2 (P. putida K2-K3), 2 (P. putida W619), 3 (P. aeruginosa H1), 4A (P. aeruginosa H3), 4B (P. putida K1) and 5 (A. tumefacines).
The reference strain P. putida KT2440 is equipped with three T6SSs
We used the strain KT2440 to perform in-depth genomic analysis. In this strain, only five T6SS-related genes, that is, the hcp genes PP2615 and PP4082 or the vgrG genes PP2614, PP3386 and PP4049 are annotated (http://pseudomonas.com/). Using bioinformatics approaches (for example, BLASTP, Ugene or SMART) we identified a large number of T6SS-related orfs (Supplementary Tables S4–S6). Most of the genes fall into three clusters that we named K1-, K2- and K3-T6SS (Figures 1b–d and Supplementary Tables S4 and S5). Several hcp and vgrG orphan genes were also found scattered on the chromosome (Figures 1b and e, and Supplementary Table S6). Phylogenetic analysis showed that the K2- and K3-T6SSs are related (group 1.2, Figure 2), whereas the K1 cluster clades separately (group 4B, Figure 2).
K2 and K3 consist of two divergently transcribed gene clusters that contain 12 of the 13 genes encoding core T6SS components (Figure 1d). The missing core gene, clpV, encodes the ATPase required for disassembling the sheath (Kapitein et al., 2013; Kube et al., 2014), which is absent in all clusters belonging to group 1.2 (data not shown). Using the ‘CoGe's Genome Evolution Analysis’ tool, we observed a synteny among the K2 and K3 clusters (Figure 1d). The identity of the corresponding proteins encoded within each of these clusters was remarkably high, ranging from 64 to 99% (Supplementary Table S5). These observations indicate that the two clusters may have arisen from a duplication event.
The K1 system is not related to K2 and K3, and belongs to the plant-related group (group 4B, Figure 2; Boyer et al., 2009). This cluster comprises two putative operons and an ‘intermediate’ region (Figure 1c). The first operon contains 15 genes, 12 of which encode T6SS core components, and was named the structural operon (Figure 1c). The last core component gene, vgrG, is located within the second operon that was therefore named the VgrG1 operon (Figure 1c). Within the structural operon we found a previously undefined orf, PP3090.1 encoding the accessory protein TagF1 (Supplementary Table S4). An ortholog of this protein was reported to function as a posttranscriptional regulator (Silverman et al., 2011). Another accessory gene encodes TagP1 (Supplementary Table S4), a TssM derivative whose C-terminal periplasmic portion carries a peptydoglycan-binding domain (pfam00691; Aschtgen et al., 2010). Finally, our analysis identified a novel T6SS feature represented by the first gene in the K1-T6SS structural operon, PP3101.1, tagX1 (Figure 1c and Supplementary Table S4). The protein encoded by this gene has no homologs or recognizable features. It has not been assigned a role in the T6SS but is exclusively present in all clusters belonging to the 4B group (that is, P. putida and Pseudomonas syringae).
The K1-T6SS is functional and anti-bacterial
Hcp release is dependent on the T6SS and is a reliable marker for assessing functionality of the system (Pukatzki et al., 2006). Therefore, we engineered P. putida strains producing an HA-tagged version of Hcp1 to assess K1-T6SS activity. TssA is a core baseplate component of the T6SS, is essential for T6SS activity (Planamente et al., 2016) and we used a tssA mutant to disable the P. putida K1-T6SS. We readily detected Hcp1 in the supernatant of wild-type cultures but not in an isogenic tssA1 mutant (Figure 3a), thus establishing that the K1-T6SS is a functional secretion machine.
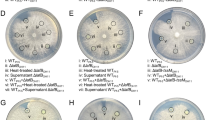
Functionality of the P. putida K1-T6SS. (a) Production and secretion of Hcp1 in the P. putida KT2440 wild type and the ΔtssA1 mutant strains. The HA-tagged Hcp1 protein was detected by western blot analysis using an anti-HA antibody. Detection of the β-subunit of the RNA polymerase (β-RNAP) was used as control. The position of the molecular size marker (in kDa) is indicated. (b) Competition assay between P. putida and a lacZ-encoding E. coli strain. Blue patches on X-gal-containing LB plates indicate E. coli survival. The top row shows the growth of E. coli, P. putida KT2440 wild-type strain and a battery of P. putida mutants in K1-T6SS genes. The bottom row shows the growth of mixed E. coli/P. putida cultures after 5 h of co-incubation.
Several characterized T6SSs have anti-bacterial activity, resulting from the injection of T6SS toxins into bacterial preys (Russell et al., 2014; Cianfanelli et al., 2016). We performed competition assays using E. coli K12 as prey and P. putida wild type or T6SS mutants as predators. The E. coli prey harbors a plasmid that confers blue color to the colony in the presence of X-gal (Figure 3b). In a mixed culture, the P. putida wild-type strain was able to annihilate E. coli, whereas mutants in any of the K1-T6SS structural genes (tssA1, tssL1, tssK1, tssG1, tssF1 or tssE1) were no longer outcompeting E. coli (Figure 3b). In contrast, mutants in the K2- or K3-T6SS clusters, P. putida ΔtssM2 and ΔtssM3, respectively, still efficiently annihilated E. coli (data not shown). We concluded that K1 is the most active KT2440 T6SS in vitro, as under the laboratory conditions used here, and that its antibacterial activity may result from the secretion of T6SS effectors.
P. putida KT2440 encodes a wealth of T6SS bacterial effectors
Genes encoding putative T6SS effectors and cognate immunity proteins (effector–immunity (EI) pairs) are often linked to hcp, vgrG genes and/or genes encoding chaperones/adaptors (Dong et al., 2013; Hachani et al., 2014; Ma et al., 2014; Liang et al., 2015; Unterweger et al., 2015). Our in-silico analyses identified a total of 10 potential EI pairs, most of them encoded in the vicinity of vgrG/hcp genes and in some cases near genes encoding Tap or EagR adaptors (Figures 1c–e and Supplementary Tables S4–S6). These EI pairs have been named Tke and Tki for Type six KT2440 effector and immunity, respectively (Figure 4a).
P. putida KT2440 T6SS effectors. (a) The domain organization of the putative effectors is shown, with PAAR motifs indicated in orange, MIX motifs in blue, Rhs domains in green, HNH nuclease motifs (Tox-HNH and Tox-SHH) in purple, colicin motifs in yellow and the Tox-61 domain in pink. Multiple sequence alignments of the PAAR (b) and MIX (c) protein motifs are represented. The KT2440 T6SS effectors identified in this work are indicated in blue. The sequence of known T6SS effectors containing these motifs was retrieved from the NCBI database (http://www.ncbi.nlm.nih.gov/Structure/cdd/cdd.shtml). Conservation logos of the motifs are indicated above the alignment. Conserved residues are highlighted according to the amino acid characteristic: hydrophobic (black), small (pink), positive (blue), negative (yellow) and polar (purple, light blue, red).
VgrG linked effectors
Downstream vgrG1 and vgrG2 in the K1 and K2 clusters, respectively, putative effector genes, tke2 and tke4, and EagR adaptor genes, eagR1a-eagR1b and eagR2, were found (Figures 1c and d). Tke2 and Tke4 proteins share a similar structure, both containing an N-terminal PAAR motif (Cascales and Cambillau, 2012; Shneider et al., 2013) and a conserved Rhs domain (Busby et al., 2013) limited by specific RVxxxxxxxxG and PxxxxDPxGL motifs (Figures 4a and b). PAAR proteins have been shown to be located at the tip of the VgrG trimer, sharpening the T6SS spike and/or creating an interface for T6SS effectors and adaptors (Whitney et al., 2015). The C-terminal region of Tke2 or Tke4 (110 and 102 amino acid long, respectively) carries a cytotoxic domain. This domain is similar in both proteins and belongs to the HNH superfamily of nucleases, for example, colicin E7 and pyocin S1 (Figures 4a and 5a; Huang and Yuan, 2007), although Tke4 domain contains a specific SHH signature (Figures 4a and 5b). Genes encoding putative effectors were also found downstream vgrG3, vgrG4 and vgrG5 (Figures 1d and e). The tke5 and tke9 genes within the K3 and vgrG4 operons, respectively, are linked to genes encoding Tap adaptors (tap3 and tap4; Figures 1d and e). No recognizable features were found in Tke5 or Tke9, except for a conserved N-terminal MIX motif considered a marker for T6SS effectors (Salomon et al., 2014; Figures 4a and c). This motif is also present in the effector-encoded downstream vgrG5, Tke10, which is predicted to be a restriction endonuclease (Figures 1e and 4c). In addition, tke5 and tke10 are linked to genes encoding a PAAR-motif (named tsp for type six paar; tsp5 and tsp10; Figures 1d, e and 4b, and Supplementary Table S6).
P. putida KT2440 T6SS nucleases. (a, b) Multiple sequence alignments of the C-terminal domains of Tke2 (a) and Tke4 (b) effectors (blue) with known proteins of the family (black). Conservation logos of the motifs HNH (a) and SHH (b) are indicated above the alignment. Conserved residues are indicated with the color code used in Figure 4. A representation of the structural model of the C-terminal domain of the Tke2 effector (magenta) superimposed on the colicin E7 structure (blue; PDB: 2JB0) is shown on the right of a. Side chains of the active site residues are shown. (c) Multiple sequence alignment of T6SS colicin effectors (blue) with known proteins of the family (black). The secondary structure prediction (ssp) for effector Tke7 is shown above the alignment. A structural alignment of the Tke7 effector model (magenta) with the colicin S4 (blue, PDB: 3FEW) is shown on the right.
Effectors encoded in proximity to hcp genes
The potential effector genes tke6, tke7 and tke8 were found within or in the vicinity of the three hcp orphan operons (hcp4, hcp5 and hcp6; Figure 1e and Supplementary Table S6). These effectors have similarities with pore-forming colicins (that is, colicin S4; Figures 4a and 5c, and Supplementary Table S6). The tke7 and tke8 genes are not genetically associated with vgrG or T6SS adaptor genes. These hcp-linked T6SS effectors could be delivered by docking into the lumen of the Hcp ring, instead of being attached at the VgrG tip, as observed with the P. aeruginosa Tse2 effector (Silverman et al., 2013). In contrast to tke7 and tke8, tke6 is not located within the hcp operon but 5 kb upstream of the hcp4 gene. Interestingly, hcp4 has a premature stop codon and might not be functional (Figure 1e and Supplementary Table S6), whereas Tke6 has an N-terminal PAAR domain (Figures 4a and b). Thus, in contrast to Tke7 and Tke8 that lack PAAR or MIX domains, the delivery of Tke6 could be mediated by a VgrG protein through a PAAR-VgrG interaction.
Orphan effectors
We found two additional potential EI pairs (tke1-tki1 and tke3-tki3) within the K1-T6SS cluster, both lacking PAAR or MIX motifs. Tke1 is an ortholog of the P. aeruginosa Tse6, which presents a C-terminal region carrying a toxic domain known as Toxin_61 (Figure 4a and Supplementary Figure S2a; Hachani et al., 2014; Whitney et al., 2014) and degrades NAD(P)(+) in target cells (Whitney et al., 2015). In case of Tke3, a Phyre2 analysis suggests that the C-terminal domain resembles the B30.2 fragment from the human protein TRIM20 (Weinert et al., 2015; Supplementary Figure S2b and Supplementary Table S4).
In summary, we identified 10 potential T6SS effectors in the KT2440 genome. Three of them Tke2, Tke4 and Tke6 have an N-terminal PAAR domain (Figures 4a and b) and are therefore considered ‘specialized’ effectors. The others are not fused to any T6SS component and their domain architecture suggests they are ‘cargo’ effectors.
Tke2/Tki2 is a P. putida K1-T6SS effector/immunity pair
We have shown that the K1 system is functional, and that the corresponding gene cluster encodes several EI pairs including Tke2/Tki2 (Figures 1c, 4a and 5a, and Supplementary Table S4). To assess the functionality of this EI pair, the tke2 and tki2 genes were cloned into compatible plasmids and transformed into E. coli K12. Expression was induced by the addition of isopropyl β-D-1-thiogalactopyranoside (tke2) or arabinose (tki2). On induction of the effector gene tke2, E. coli growth was significantly impaired (Figures 6a and b) but growth could readily be rescued on co-expression of the putative Tki2 immunity protein (Figures 6a and b). This suggests that Tke2/Tki2 is a genuine EI pair.
Toxicity and secretion of the Tke2 effector. (a) The growth of E. coli K12 cells harboring the pTke2-CT and pTki2 plasmids containing the C-terminal toxin domain of the tke2 effector and the tki2 immunity genes, respectively, was determined by measuring the OD at 600 nm. At time zero, either 1 mM isopropyl β-D-1-thiogalactopyranoside (IPTG) and/or 0.02% (w/v) arabinose were added to the LB medium, to induce expression of the tke2-CT or/and tki2 genes, respectively. (b) Western blot analyses using an anti-V5 or anti-HA monoclonal antibody to detect the Tke2-CT-V5 or Tki2-HA-tagged proteins. Proteins were prepared from E. coli K12 cells grown during 10 h in presence (+) or absence (−) of 1 mM IPTG and/or 0.02% (w/v) arabinose. (c) The indicated P. putida KT2440 strains bearing a tke2-V5-tagged gene were grown in tryptone soya broth (TSB) medium for 5 h. Tke2-V5 was detected in the whole cell and supernatant fractions using a monoclonal anti-V5 antibody. Detection of the β-subunit of the RNA polymerase (β-RNAP) was used as control. The position of the molecular size marker (in kDa) is indicated.
We assessed whether Tke2 is secreted in a K1-T6SS-dependent manner. The corresponding gene was replaced on the KT2440 chromosome with a version encoding a C-terminally V5-tagged protein. However, Tke2-V5 production was only weakly detected when using this strain (Supplementary Figure S3). In contrast to bacterial killing, which is a highly sensitive assay, detection of secreted T6SS toxins by western blot may need higher level of T6SS expression (Cianfanelli et al., 2016). It has been described in other bacteria that several global regulators are involved in T6SS expression, including the alternative sigma factor RpoN (Bernard et al., 2010, 2011; Sana et al., 2013). The tke2-V5 chimeric gene was introduced into an rpoN mutant and in this strain Tke2 production was considerably increased as compared with the wild-type P. putida (Supplementary Figure S3). We thus used this genetic background to analyze Tke2 secretion. Tke2 was produced in both the rpoN strain and the isogenic T6SS mutant (rpoNΔtssA1), but was only found in the supernatant of the strain with an intact T6SS (Figure 6c). Our results show that Tke2 is an effector of the K1-T6SS and its activity is antagonized by the Tki2 immunity protein.
P. putida outcompetes plant pathogens in a T6SS-dependent manner
P. putida is an efficient biocontrol agent (Amer and Utkhede, 2000; Validov et al., 2007) and we hypothesized that it uses the T6SS to kill ecologically relevant competitors. To test this we selected four plant pathogens, P. syringae, X. campestris, Pectobacterium carotovorum and Agrobacterium tumefaciens, which are leading causes of deadly diseases in several economically important crops (Mansfield et al., 2012). The various T6SSs are likely to be differentially expressed in vitro, in vivo, in planta or in the presence of different competitors (Ma et al., 2014). To assess whether the T6SS in general is required for outcompeting plant pathogens and thus involve in plant protection we used a triple T6SS mutant (ΔtssA1ΔtssM2ΔtssM3, also named ΔT6SS), so that none of the K1, K2 or K3 system is at play. First, a competition assay between KT2440 or the triple mutant and the phytopathogens was performed. The P. putida wild-type strain caused a 10-fold decrease in survival of A. tumefaciens and P. caratovorum, and a 1000-fold decrease in the survival of X. campestris and P. syringae (Figure 7). The P. putida T6SS mutant had barely any impact on the survival of any of these bacteria (Figure 7). Our results indicate that KT2440 outcompetes all challenged phytopathogens in a T6SS-dependent manner and suggest a role for this secretion system in biocontrol.
Bactericidal activity of P. putida KT2440 against a panel of phytopathogens. X. campestris, A. tumefaciens, P. carotovorum and P. syringae pv. tomato strains harbor the pRL662-gfp plasmid that confers gentamycin resistance. The P. putida KT2440 wild type (WT) and its isogenic ΔtssA1ΔtssM2ΔtssM3 triple mutant (ΔT6SS) were co-incubated with the phytopathogens for 24 h. Colony-forming unit (CFU) quantifications were performed on gentamycin selection. The average±s.d. from at least three biological replicates is plotted.
T6SS-active P. putida protects plants from pathogen’s attack
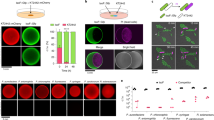
To assess the ability of P. putida to kill phytopathogens in an ecologically relevant set-up, we developed an in planta competition assay. We selected X. campestris as the pathogen and N. benthamiana as the plant model. Leaves were co-infected with X. campestris and either P. putida wild type or the isogenic ΔT6SS mutant. X. campestris was tagged with a green fluorescent protein to monitor in situ colonization. X. campestris-induced halos of necrosis on the leaves were observed 5 days post infection, whereas inoculation with P. putida resulted in healthy-looking leaves (Figure 8a). Remarkably, co-infiltration of X. campestris and P. putida wild-type strain considerably reduced the necrotic areas produced by X. campestris (circled in Figure 8b lower panel). This is not observable with the P. putida ΔT6SS mutant and we concluded that interference with X. campestris colonization is T6SS dependent (Figure 8b). The protection conferred by P. putida is due to reduced survival of X. campestris in the leaves (~2.5-fold reduction), as qualitatively observed by fluorescence microscopy (Figure 8b upper panel) and quantitatively measured by colony-forming unit counting (Figure 8c). Our results show that P. putida outcompetes X. campestris during plant colonization and this process involves the bactericidal properties of the T6SS.
In planta competition assay between the biocontrol strain P. putida KT2440 and the phytopathogen X. campestris. (a) Leaves of N. benthamiana 24 h (upper panel) and 5 days (lower panel) after being infiltrated with X. campestris (pRL662-gfp; expressing a plasmid-encoded green fluorescence protein), the P. putida KT2440 wild type (WT), or its isogenic ΔtssA1ΔtssM2ΔtssM3 triple mutant (ΔT6SS). (b) Leaves of N. benthamiana 24 h (upper panel) and 5 days (lower panel) after co-infiltration of X. campestris (pRL662-gfp) with the indicated P. putida strain. In upper panel a and b, the leaves were visualized by fluorescence microscopy using a Leica M205FA stereomicroscope. The necrotic areas resulting from X. campestris infection are marked. The deep brown zone of necrosis is spread on a large portion of the leave (right panel), while such spread is far more restricted when the phytopathogen is co-inoculated with a T6SS positive P. putida strain (left panel). (c) Quantification of X. campestris (pRL662-gfp) colony-forming units (CFUs) recovered from N. benthamiana leaves after 24 h of co-infiltration with the indicated P. putida strain. X. campestris CFU were quantified after gentamycin (Gm) selection. Graphs represent mean +s.d., of at least five biological replicates with two technical replicates per experiment, statistical significance is indicated t-test P<0.001.
Discussion
The T6SS was discovered in the bacterial pathogens V. cholerae (Pukatzki et al., 2006) and P. aeruginosa (Mougous et al., 2006). Since then, an increasing number of studies has provided details on the function and structure of this original bacterial secretion system (Russell et al., 2014; Zoued et al., 2014; Cianfanelli, et al., 2016; Hachani et al., 2016). However, although the presence of the T6SS in non-pathogenic strains is evident (Boyer et al., 2009), little work has been done to understand its relevance in this category of bacteria (Bladergroen et al., 2003; Mougous et al., 2006; Pukatzki et al., 2006; Aschtgen et al., 2008).
Phylogeny and genetic structure of the P. putida T6SS clusters
In this study we have identified a total of 66 T6SS clusters among P. putida strains, which suggests that this secretion machine has an important role in P. putida physiology and fitness. The P. putida T6SS clusters clade within three phylogenetic groups, group 1.2, 2 or 4B (Figure 2). Remarkably, P. putida is the only Pseudomonas species encoding T6SSs from group 1.2, whereas T6SSs from group 4B are only present in P. putida and P. syringae (Barret et al., 2011). The P. putida KT2440 strain contains two clusters from group 1.2 (K2 and K3) and one cluster from group 4B (K1). The K2-T6SS cluster contains two orfs, vgrG2 and tssC2, which present premature stop codons (Supplementary Table S5), implying that this system is not functional. Prematurely interrupted T6SS genes have been identified in functional T6SSs of Citrobacter rodentium and Yersinia pseudotuberculosis (Gueguen et al., 2014). In these cases, a transcriptional frameshifting caused by a poly-A tract allows the production of functional TssM variants (Gueguen et al., 2014). However, this is unlikely to be the case in KT2440, as poly-A tracts are not found either in tssC2 or in vgrG2. Alternatively, related VgrGs (that is, VgrG3, VgrG4 and VgrG5; Supplementary Figure S4) and TssC proteins (that is, TssC3; Supplementary Table S5) could be shared between different T6SSs.
The K2 and K3 clusters do not encode a ClpV protein, the ATPase responsible for disassembling the T6SS sheath. Yet, orphan clpV genes can be used. There are three Clp ATPase-encoding genes in the KT2440 genome (that is, PP0625, PP3316 and PP4008), but none encodes a protein from the ClpV family (Supplementary Figure S5). They are ClpA and ClpB members, which are phylogenetically distant from ClpV (Schlieker et al., 2005). Alternatively, the ClpV1 component within the K1 cluster could be shared between the systems but possibly a ClpV component may not be necessary for the function of the group 1.2 T6SS, as some P. putida strains (that is, S12, B001, SJ3 and S610) exclusively contain a group 1.2 cluster (Supplementary Table S3). In fact, functional T6SSs lacking the clpV gene have been identified in other bacteria (Chow and Mazmanian, 2010; Bröms et al., 2012). Furthermore, the clpV gene of V. cholerae can be deleted without a total loss of T6SS function (Zheng et al., 2011). After all, other nanomachines structurally comparable to the T6SS such as the contractile-tailed phages or R-type pyocins, do not use a ClpV homolog for their function. Instead, recently discovered phage-like protein translocation structures are encoded within gene clusters that also carry a clpV homolog (Kube and Wendler, 2015). This type of structure may have evolved divergently with some of the T6SS subgroups and acquired ClpV from ancestral systems.
Antibacterial activity of the P. putida KT2440 T6SS
The main role of the T6SS is to inject effectors into eukaryotic or prokaryotic prey cells (Alcoforado Diniz et al., 2015; Hachani et al., 2016). We identified an impressive battery of 10 potential T6SS effectors in P. putida KT2440. This is not unique but suggests that P. putida is primed to fight a wide range of competing organisms. At least three EI pairs are encoded within the K1-T6SS cluster (that is, tke1-tki1, tke2-tki2 and tke3-tki3), which belongs to the uncharacterized plant-related group (group 4B, Figure 2). A remarkable characteristic of the system is the presence of a conserved accessory gene, tagX, systematically absent from other T6SS groups and which is a hallmark for group 4B systems. Here we show that suitable preys for the K1-T6SS are bacterial cells, and that the Tke2 toxin contributes to the antibacterial activity. Tke2 contains a canonical Rhs-effector domain organization, which includes an N-terminal PAAR motif, a central domain of conserved Rhs-repeats and a C-terminal toxic domain. Although the function of the Rhs domain is still unknown, it has been suggested that it forms a shell structure that encapsulates the C-terminal region of effectors (Busby et al., 2013; Supplementary Figure S6). Furthermore, a specific adaptor named EagR (after ‘effector-associated gene’) that contains the DUF1795 domain, has been involved in the secretion of PAAR/Rhs effectors (Alcoforado Diniz and Coulthurst, 2015). Two different proteins containing DUF1795 domains are encoded immediately upstream tke2 (eagR1a and eagR1b, Figure 1c). Although the function of these adaptors has not been analyzed yet, it is possible that both function together to assist Tke2 secretion. The recurrent association between PAAR/Rhs effectors and EagR adaptors is furthermore confirmed by the association of tke4, encoding another P. putida PAAR/Rhs effector (Figures 1c and 4a, and Supplementary Figure S6), with an eagR gene (eagR2).
Biocontrol properties of the P. putida T6SS
It is becoming increasingly obvious that the antimicrobial properties of the T6SS could be instrumental for the control of polymicrobial populations in excluding foes from natural and ecologically relevant environments. For instance, a clear correlation between activation of T6SS, enhanced fitness and subsequent antagonism against other bacteria has been observed with Vibrio parahaemolyticus in marine niches (Salomon et al., 2013). This suggested that T6SSs are key for survival and persistence of specialized species in specific habitats. In the lungs of cystic fibrosis patients, P. aeruginosa can persist for years, while the diversity of species that primarily colonizes this environment decreases over time (Marshall et al., 2015). P. aeruginosa isolates from cystic fibrosis patients have highly active T6SSs (Mougous et al., 2006; Moscoso et al., 2011), which suggests that T6SSs contribute to the colonization advantage of P. aeruginosa over other species. In agreement with these observations, the T6SS has been proposed to be crucial in the establishment/evolution of the gut microbiome (Russell, et al., 2014; Cianfanelli, et al., 2016). Half of the human-associated Bacteroidetes, the dominant phyla in the human gut, not only encode T6SSs (Coyne et al., 2016) and possess a wide range of T6SS effectors (Chatzidaki-Livanis et al., 2016) but accumulate immunity genes against other T6SS effectors (Wexler et al., 2016). This strongly supports that T6SS is a selective mechanism involved in the establishment of gut communities. These remarkable properties of the T6SS are obviously useful in the development of biocontrol strains. The T6SS was originally discovered in Rhizobium leguminosarum and involved in pea nodulation (Bladergroen et al., 2003), but barely any studies have demonstrated the potential that such system may have in the context of the plant microbiome. A parallel can be made between the gut and the rhizosphere, as both are eukaryotic-based environments hosting a symbiotic relationship with a complex microbial community (Stone, 2016). Both animals and plants depend on their microbiome to protect themselves against pathogens and to help assimilate necessary nutrients (Carmody et al., 2015; Haney and Ausubel, 2015; Haney et al., 2015). As a defence strategy, many plant species promote the development of a specific microbiome in the rhizosphere, which has antagonistic activity against soil-borne pathogens (Cook et al., 1995; Weller et al., 2002; Lebeis et al., 2015). Although the mechanisms for pathogen suppression are not completely understood, they include the production of bioactive metabolites such as antibiotics, bacteriocins and siderophores (Weller, 2007). However, these mechanisms fail to account for the full level of protection conferred by the biocontrol organism (Matilla et al., 2010). Here we report for the first time that the T6SS might be a primary mechanism for phytopathogen control. Indeed, we demonstrate that the crop protection agent P. putida KT2440 readily outcompetes a panel of economically important phytopathogens and that the efficient destruction of the pest is mostly T6SS dependent. This property can likely be transferred to the field, as this effect was observed in vitro but also in vivo by demonstrating that P. putida protects plant leaves from the deleterious effect of X. campestris.
In our study we have used a laboratory setup and further trials in crop plants are needed so that in-depth investigation of the impact of KT2440 in the rhizosphere can be assessed. Nevertheless, our finding shows that the T6SS can be used by environmental strains to protect plants from the attack of bacterial pathogens and can thus be considered as a plant health warden. This opens new possibilities in the selection of biocontrol agents used for biotechnological applications. Noticeably, the poor specificity of the T6SS (Hood et al., 2010) may allow such biocontrol organism to also fight eukaryotic pathogens belonging to different kingdoms including nematodes and fungi.
References
Alcoforado Diniz J, Coulthurst SJ . (2015). Intraspecies competition in serratia marcescens is mediated by type VI-secreted Rhs effectors and a conserved effector-associated accessory protein. J Bacteriol 197: 2350–2360.
Alcoforado Diniz J, Liu Y-CC, Coulthurst SJ . (2015). Molecular weaponry: diverse effectors delivered by the Type VI secretion system. Cell Microbiol 17: 1742–1751.
Amer GA, Utkhede RS . (2000). Development of formulations of biological agents for management of root rot of lettuce and cucumber. Can J Microbiol 46: 809–816.
Aschtgen M-SS, Bernard CS, De Bentzmann S, Lloubès R, Cascales E . (2008). SciN is an outer membrane lipoprotein required for type VI secretion in enteroaggregative Escherichia coli. J Bacteriol 190: 7523–7531.
Aschtgen M-SS, Thomas MS, Cascales E . (2010). Anchoring the type VI secretion system to the peptidoglycan: TssL, TagL, TagP... what else? Virulence 1: 535–540.
Barret M, Egan F, Fargier E, Morrissey JP, O’Gara F . (2011). Genomic analysis of the type VI secretion systems in Pseudomonas spp.: novel clusters and putative effectors uncovered. Microbiology 157: 1726–1739.
Basler M, Ho BT, Mekalanos JJ . (2013). Tit-for-tat: type VI secretion system counterattack during bacterial cell-cell interactions. Cell 152: 884–894.
Basler M, Mekalanos JJ . (2012). Type 6 secretion dynamics within and between bacterial cells. Science 337: 815.
Bernard CS, Brunet YR, Gavioli M, Lloubès R, Cascales E . (2011). Regulation of type VI secretion gene clusters by sigma54 and cognate enhancer binding proteins. J Bacteriol 193: 2158–2167.
Bernard CS, Brunet YR, Gueguen E, Cascales E . (2010). Nooks and crannies in type VI secretion regulation. J Bacteriol 192: 3850–3860.
Bladergroen MR, Badelt K, Spaink HP . (2003). Infection-blocking genes of a symbiotic Rhizobium leguminosarum strain that are involved in temperature-dependent protein secretion. Mol Plant Microbe Interact 16: 53–64.
Boratyn GM, Camacho C, Cooper PS, Coulouris G, Fong A, Ma N et al. (2013). BLAST: a more efficient report with usability improvements. Nucleic Acids Res 41: W29–W33.
Boyer F, Fichant G, Berthod J, Vandenbrouck Y, Attree I . (2009). Dissecting the bacterial type VI secretion system by a genome wide in silico analysis: what can be learned from available microbial genomic resources? BMC Genomics 10: 104.
Bröms JE, Meyer L, Sun K, Lavander M, Sjöstedt A . (2012). Unique substrates secreted by the type VI secretion system of Francisella tularensis during intramacrophage infection. PLoS ONE 7: e50473.
Burtnick MN, Brett PJ, Harding SV, Ngugi SA, Ribot WJ, Chantratita N et al. (2011). The cluster 1 type VI secretion system is a major virulence determinant in Burkholderia pseudomallei. Infect Immun 79: 1512–1525.
Busby JN, Panjikar S, Landsberg MJ, Hurst MR, Lott JS . (2013). The BC component of ABC toxins is an RHS-repeat-containing protein encapsulation device. Nature 501: 547–550.
Carmody RN, Gerber GK, Luevano JM, Gatti DM, Somes L, Svenson KL et al. (2015). Diet dominates host genotype in shaping the murine gut microbiota. Cell Host Microbe 17: 72–84.
Cascales E, Cambillau C . (2012). Structural biology of type VI secretion systems. Philos Trans R Soc Lond B Biol Sci 367: 1102–1111.
Chatzidaki-Livanis M, Geva-Zatorsky N, Comstock LE . (2016). Bacteroides fragilis type VI secretion systems use novel effector and immunity proteins to antagonize human gut Bacteroidales species. Proc Natl Acad Sci USA 113: 3627–3632.
Choi K-HH, Kumar A, Schweizer HP . (2006). A 10-min method for preparation of highly electrocompetent Pseudomonas aeruginosa cells: application for DNA fragment transfer between chromosomes and plasmid transformation. J Microbiol Methods 64: 391–397.
Chow J, Mazmanian SK . (2010). A pathobiont of the microbiota balances host colonization and intestinal inflammation. Cell Host Microbe 7: 265–276.
Cianfanelli FR, Alcoforado Diniz J, Guo M, De Cesare V, Trost M, Coulthurst SJ . (2016). VgrG and PAAR proteins define distinct versions of a functional type VI secretion system. PLoS Pathog 12: e1005735.
Cianfanelli FR, Monlezun L, Coulthurst SJ . (2016). Aim, load, fire: the type VI secretion system, a bacterial nanoweapon. Trends Microbiol 24: 51–62.
Cook RJ, Thomashow LS, Weller DM, Fujimoto D, Mazzola M, Bangera G et al. (1995). Molecular mechanisms of defense by rhizobacteria against root disease. Proc Natl Acad Sci USA 92: 4197–4201.
Coyne MJ, Roelofs KG, Comstock LE . (2016). Type VI secretion systems of human gut Bacteroidales segregate into three genetic architectures, two of which are contained on mobile genetic elements. BMC Genomics 17: 58.
de Pace F, Nakazato G, Pacheco A, de Paiva JB, Sperandio V, da Silveira WD . (2010). The type VI secretion system plays a role in type 1 fimbria expression and pathogenesis of an avian pathogenic Escherichia coli strain. Infect Immun 78: 4990–4998.
Dong TG, Ho BT, Yoder-Himes DR, Mekalanos JJ . (2013). Identification of T6SS-dependent effector and immunity proteins by Tn-seq in Vibrio cholerae. Proc Natl Acad Sci USA 110: 2623–2628.
Durand E, Cambillau C, Cascales E, Journet L . (2014). VgrG, Tae, Tle, and beyond: the versatile arsenal of type VI secretion effectors. Trends Microbiol 22: 498–507.
Durand E, Nguyen VS, Zoued A, Logger L, Péhau-Arnaudet G, Aschtgen M-SS et al. (2015). Biogenesis and structure of a type VI secretion membrane core complex. Nature 523: 555–560.
Espinosa-Urgel M, Salido A, Ramos JL . (2000). Genetic analysis of functions involved in adhesion of Pseudomonas putida to seeds. J Bacteriol 182: 2363–2369.
Filloux A . (2011). Protein secretion systems in Pseudomonas aeruginosa: an essay on diversity, evolution, and function. Front Microbiol 2: 155.
Filloux A, Freemont P . (2016). Structural biology: baseplates in contractile machines. Nat Microbiol 1: 16104.
Finn RD, Coggill P, Eberhardt RY, Eddy SR, Mistry J, Mitchell AL et al. (2016). The Pfam protein families database: towards a more sustainable future. Nucleic Acids Res 44: D279–D285.
Gomi M, Sonoyama M, Mitaku S . (2004). High performance system for signal peptide prediction: SOSUIsignal. Chem Bio Info J 4: 142–147.
Gotfredsen M, Gerdes K . (1998). The Escherichia coli relBE genes belong to a new toxin-antitoxin gene family. Mol Microbiol 29: 1065–1076.
Gueguen E, Wills NM, Atkins JF, Cascales E . (2014). Transcriptional frameshifting rescues Citrobacter rodentium type VI secretion by the production of two length variants from the prematurely interrupted tssM gene. PLoS Genet 10: e1004869.
Guzman LM, Belin D, Carson MJ, Beckwith J . (1995). Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J Bacteriol 177: 4121–4130.
Hachani A, Allsopp LP, Oduko Y, Filloux A . (2014). The VgrG proteins are ‘A la carte’ delivery systems for bacterial type VI effectors. J Biol Chem 289: 17872–17884.
Hachani A, Lossi N, Hamilton A, Jones C, Bleves S, Albesa-Jové D et al. (2011). Type VI secretion system in Pseudomonas aeruginosa secretion and multimerization of VgrG proteins. J Biol Chem 286: 12317–12327.
Hachani A, Lossi NS, Filloux A . (2013). A visual assay to monitor T6SS-mediated bacterial competition. J Vis Exp 73: e50103.
Hachani A, Wood TE, Filloux A . (2016). Type VI secretion and anti-host effectors. Curr Opin Microbiol 29: 81–93.
Haney CH, Ausubel FM . (2015). MICROBIOME. Plant microbiome blueprints. Science 349: 788–789.
Haney CH, Samuel BS, Bush J, Ausubel FM . (2015). Associations with rhizosphere bacteria can confer an adaptive advantage to plants. Nat Plants 1: 15051.
Ho B, Dong T, Mekalanos J . (2013). A view to a kill: the bacterial type VI secretion system. Cell Host Microbe 15: 9–21.
Hoang TT, Kutchma AJ, Becher A, Schweizer HP . (2000). Integration-proficient plasmids for Pseudomonas aeruginosa: site-specific integration and use for engineering of reporter and expression strains. Plasmid 43: 59–72.
Hood R, Singh P, Hsu F, Güvener T, Carl M, Trinidad R et al. (2010). A type VI secretion system of Pseudomonas aeruginosa targets a toxin to bacteria. Cell Host Microbe 7: 25–37.
Huang H, Yuan HS . (2007). The conserved asparagine in the HNH motif serves an important structural role in metal finger endonucleases. J Mol Biol 368: 812–821.
Imai K, Asakawa N, Tsuji T, Akazawa F, Ino A, Sonoyama M et al. (2008). SOSUI-GramN: high performance prediction for sub-cellular localization of proteins in gram-negative bacteria. Bioinformation 2: 417–421.
Kaniga K, Delor I, Cornelis GR . (1991). A wide-host-range suicide vector for improving reverse genetics in gram-negative bacteria: inactivation of the blaA gene of Yersinia enterocolitica. Gene 109: 137–141.
Kapitein N, Bönemann G, Pietrosiuk A, Seyffer F, Hausser I, Locker JK et al. (2013). ClpV recycles VipA/VipB tubules and prevents non-productive tubule formation to ensure efficient type VI protein secretion. Mol Microbiol 87: 1013–1028.
Katzen F, Ferreiro DU, Oddo CG, Ielmini MV, Becker A, Pühler A et al. (1998). Xanthomonas campestris pv. campestris gum mutants: effects on xanthan biosynthesis and plant virulence. J Bacteriol 180: 1607–1617.
Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJ . (2015). The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protoc 10: 845–858.
Krogh A, Larsson B, von Heijne G, Sonnhammer EL . (2001). Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol 305: 567–580.
Kube S, Kapitein N, Zimniak T, Herzog F, Mogk A, Wendler P . (2014). Structure of the VipA/B type VI secretion complex suggests a contraction-state-specific recycling mechanism. Cell Rep 8: 20–30.
Kube S, Wendler P . (2015). Structural comparison of contractile nanomachines. AIMS Biophys 2: 88–115.
Kudryashev M, Wang RY, Brackmann M, Scherer S, Maier T, Baker D et al. (2015). Structure of the type VI secretion system contractile sheath. Cell 160: 952–962.
Lebeis SL, Paredes SH, Lundberg DS, Breakfield N, Gehring J, McDonald M et al. (2015). PLANT MICROBIOME. Salicylic acid modulates colonization of the root microbiome by specific bacterial taxa. Science 349: 860–864.
Leiman PG, Basler M, Ramagopal UA, Bonanno JB, Sauder JM, Pukatzki S et al. (2009). Type VI secretion apparatus and phage tail-associated protein complexes share a common evolutionary origin. Proc Natl Acad Sci USA 106: 4154–4159.
Letunic I, Doerks T, Bork P . (2015). SMART: recent updates, new developments and status in 2015. Nucleic Acids Res 43: D257–D260.
Liang X, Moore R, Wilton M, Wong MJ, Lam L, Dong TG . (2015). Identification of divergent type VI secretion effectors using a conserved chaperone domain. Proc Natl Acad Sci USA 112: 9106–9111.
Lin J-SS, Ma L-SS, Lai E-MM . (2013). Systematic dissection of the agrobacterium type VI secretion system reveals machinery and secreted components for subcomplex formation. PLoS One 8: e67647.
Lyons E, Freeling M . (2008). How to usefully compare homologous plant genes and chromosomes as DNA sequences. Plant J 53: 661–673.
Ma AT, Mekalanos JJ . (2010). In vivo actin cross-linking induced by Vibrio cholerae type VI secretion system is associated with intestinal inflammation. Proc Natl Acad Sci USA 107: 4365–4370.
Ma L-SS, Hachani A, Lin J-SS, Filloux A, Lai E-MM . (2014). Agrobacterium tumefaciens deploys a superfamily of type VI secretion DNase effectors as weapons for interbacterial competition in planta. Cell Host Microbe 16: 94–104.
Mansfield J, Genin S, Magori S, Citovsky V, Sriariyanum M, Ronald P et al. (2012). Top 10 plant pathogenic bacteria in molecular plant pathology. Mol Plant Pathol 13: 614–629.
Marchi M, Boutin M, Gazengel K, Rispe C, Gauthier J-PP, Guillerm-Erckelboudt A-YY et al. (2013). Genomic analysis of the biocontrol strain Pseudomonas fluorescens Pf29Arp with evidence of T3SS and T6SS gene expression on plant roots. Environ Microbiol Rep 5: 393–403.
Marshall B, Elbert A, Petren K, Rizvi S, Fink A, Ostrenga J et al. (2015) Cystic Fibrosis Foundation Patient Registry 2014 Annual Data Report. 2015 Cystic Fibrosis Foundation: Bethesda, Maryland, USA.
Matilla M, Ramos J, Bakker P, Doornbos R, Badri D, Vivanco J et al. (2010). Pseudomonas putida KT2440 causes induced systemic resistance and changes in Arabidopsis root exudation. Environ Microbiol Rep 2: 381–388.
Miyata ST, Kitaoka M, Brooks TM, McAuley SB, Pukatzki S . (2011). Vibrio cholerae requires the type VI secretion system virulence factor VasX to kill Dictyostelium discoideum. Infect Immun 79: 2941–2949.
Molina L, Ramos C, Duque E, Ronchel MC, Garcia JM, Wyke L et al. (2000). Survival of Pseudomonas putida KT2440 in soil and in the rhizosphere of plants under greenhouse and environmental conditions. Soil Biol Biochem 32: 315–321.
Moscoso JA, Mikkelsen H, Heeb S, Williams P, Filloux A . (2011). The Pseudomonas aeruginosa sensor RetS switches type III and type VI secretion via c-di-GMP signalling. Environ Microbiol 13: 3128–3138.
Mougous JD, Cuff ME, Raunser S, Shen A, Zhou M, Gifford CA et al. (2006). A virulence locus of Pseudomonas aeruginosa encodes a protein secretion apparatus. Science 312: 1526–1530.
Murdoch SL, Trunk K, English G, Fritsch MJ, Pourkarimi E, Coulthurst SJ . (2011). The opportunistic pathogen Serratia marcescens utilizes type VI secretion to target bacterial competitors. J Bacteriol 193: 6057–6069.
Okonechnikov K, Golosova O, Fursov M . (2012). Unipro UGENE: a unified bioinformatics toolkit. Bioinformatics 28: 1166–1167.
Petersen TN, Brunak S, von Heijne G, Nielsen H . (2011). SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods 8: 785–786.
Planamente S, Salih O, Manoli E, Albesa-Jové D, Freemont PS, Filloux A . (2016). TssA forms a gp6-like ring attached to the type VI secretion sheath. EMBO J 35: 1613–1627.
Pukatzki S, Ma AT, Sturtevant D, Krastins B, Sarracino D, Nelson WC et al. (2006). Identification of a conserved bacterial protein secretion system in Vibrio cholerae using the Dictyostelium host model system. Proc Natl Acad Sci USA 103: 1528–1533.
Ramos-Gonzalez MI, Duque E, Ramos JL . (1991). Conjugational transfer of recombinant DNA in cultures and in soils: host range of Pseudomonas putida TOL plasmids. Appl Environ Microbiol 57: 3020–3027.
Rosales-Reyes R, Skeldon AM, Aubert DF, Valvano MA . (2012). The type VI secretion system of Burkholderia cenocepacia affects multiple Rho family GTPases disrupting the actin cytoskeleton and the assembly of NADPH oxidase complex in macrophages. Cell Microbiol 14: 255–273.
Russell AB, Peterson SB, Mougous JD . (2014). Type VI secretion system effectors: poisons with a purpose. Nat Rev Microbiol 12: 137–148.
Russell AB, Wexler AG, Harding BN, Whitney JC, Bohn AJ, Goo YA et al. (2014). A type VI secretion-related pathway in Bacteroidetes mediates interbacterial antagonism. Cell Host Microbe 16: 227–236.
Salomon D, Gonzalez H, Updegraff BL, Orth K . (2013). Vibrio parahaemolyticus type VI secretion system 1 is activated in marine conditions to target bacteria, and is differentially regulated from system 2. PLoS One 8: e61086.
Salomon D, Kinch LN, Trudgian DC, Guo X, Klimko JA, Grishin NV et al. (2014). Marker for type VI secretion system effectors. Proc Natl Acad Sci USA 111: 9271–9276.
Sambrook J, Maniatis T, Fritsch EF . (1989) Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory: Cold Spring Harbor, NY.
Sana TG, Soscia C, Tonglet CMM, Garvis S, Bleves S . (2013). Divergent control of two type VI secretion systems by RpoN in Pseudomonas aeruginosa. PLoS One 8: e76030.
Schlieker C, Zentgraf H, Dersch P, Mogk A . (2005). ClpV, a unique Hsp100/Clp member of pathogenic proteobacteria. Biol Chem 386: 1115–1127.
Shneider MM, Buth SA, Ho BT, Basler M, Mekalanos JJ, Leiman PG . (2013). PAAR-repeat proteins sharpen and diversify the type VI secretion system spike. Nature 500: 350–353.
Silverman JM, Agnello DM, Zheng H, Andrews BT, Li M, Catalano CE et al. (2013). Haemolysin coregulated protein is an exported receptor and chaperone of type VI secretion substrates. Mol Cell 51: 584–593.
Silverman JM, Austin LS, Hsu F, Hicks KG, Hood RD, Mougous JD . (2011). Separate inputs modulate phosphorylation-dependent and -independent type VI secretion activation. Mol Microbiol 82: 1277–1290.
Stone M . (2016). Root and gut microbiomes are strikingly similar. Microbe 11: 107–110.
Suarez G, Sierra JC, Sha J, Wang S, Erova TE, Fadl AA et al. (2008). Molecular characterization of a functional type VI secretion system from a clinical isolate of Aeromonas hydrophila. Microb Pathog 44: 344–361.
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S . (2013). MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30: 2725–2729.
Unterweger D, Kostiuk B, Ötjengerdes R, Wilton A, Diaz-Satizabal L, Pukatzki S . (2015). Chimeric adaptor proteins translocate diverse type VI secretion system effectors in Vibrio cholerae. EMBO J 34: 2198–2210.
Validov S, Kamilova F, Qi S, Stephan D, Wang JJ, Makarova N et al. (2007). Selection of bacteria able to control Fusarium oxysporum f. sp. radicis-lycopersici in stonewool substrate. J Appl Microbiol 102: 461–471.
Vasseur P, Vallet-Gely I, Soscia C, Genin S, Filloux A . (2005). The pel genes of the Pseudomonas aeruginosa PAK strain are involved at early and late stages of biofilm formation. Microbiology 151: 985–997.
Weinert C, Morger D, Djekic A, Grütter MG, Mittl PR . (2015). Crystal structure of TRIM20 C-terminal coiled-coil/B30.2 fragment: implications for the recognition of higher order oligomers. Sci Rep 5: 10819.
Weller DM . (2007). Pseudomonas biocontrol agents of soilborne pathogens: looking back over 30 years. Phytopathology 97: 250–256.
Weller DM, Raaijmakers JM, Gardener BB, Thomashow LS . (2002). Microbial populations responsible for specific soil suppressiveness to plant pathogens. Annu Rev Phytopathol 40: 309–348.
Wexler AG, Bao Y, Whitney JC, Bobay L-MM, Xavier JB, Schofield WB et al. (2016). Human symbionts inject and neutralize antibacterial toxins to persist in the gut. Proc Natl Acad Sci USA 113: 3639–3644.
Whitney JC, Beck CM, Goo YA, Russell AB, Harding BN, De Leon JA et al. (2014). Genetically distinct pathways guide effector export through the type VI secretion system. Mol Microbiol 92: 529–542.
Whitney JC, Quentin D, Sawai S, LeRoux M, Harding BN, Ledvina HE et al. (2015). An interbacterial NAD(P)(+) glycohydrolase toxin requires elongation factor Tu for delivery to target cells. Cell 163: 607–619.
Winsor GL, Griffiths EJ, Lo R, Dhillon BK, Shay JA, Brinkman FS . (2016). Enhanced annotations and features for comparing thousands of Pseudomonas genomes in the Pseudomonas genome database. Nucleic Acids Res 44: D646–D653.
Yu NY, Wagner JR, Laird MR, Melli G, Rey S, Lo R et al. (2010). PSORTb 3.0: improved protein subcellular localization prediction with refined localization subcategories and predictive capabilities for all prokaryotes. Bioinformatics 26: 1608–1615.
Zheng J, Ho B, Mekalanos JJ . (2011). Genetic analysis of anti-amoebae and anti-bacterial activities of the type VI secretion system in Vibrio cholerae. PLoS One 6: e23876.
Zoued A, Brunet YR, Durand E, Aschtgen M-SS, Logger L, Douzi B et al. (2014). Architecture and assembly of the Type VI secretion system. Biochim Biophys Acta 1843: 1664–1673.
Acknowledgements
We thank Milagros Lopez (IVIA, Spain) for providing X. campestris and P. carotovorum, Martin Buck (Imperial College London, UK) for P. syringae and Ehr Min Lai (Academia Sinica, Taiwan) for A. tumefaciens and the pRL662-gpf. We thank Tom Wood for kindly providing the T6SS scheme shown in Figure 1a. PB is supported by the Spanish Ministry of Economy through Juan de la Cierva grant (JCI-2010-06615), by the Andalusian Knowledge Agency through a Talent Hub grant (TAHUB-010) and by an EMBO short-term fellowship. MAL is supported by the Spanish Ministry of Economy through a Ramon&Cajal grant (RYC2011-08874). AF is supported by a BBSRC grant (BB/N002539/1). LA is supported by a BBSRC grant (BB/N002539/1) and a Marie curie Fellowship (PIIF-GA-2012-328261).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on The ISME Journal website
Supplementary information
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Bernal, P., Allsopp, L., Filloux, A. et al. The Pseudomonas putida T6SS is a plant warden against phytopathogens. ISME J 11, 972–987 (2017). https://doi.org/10.1038/ismej.2016.169
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ismej.2016.169
This article is cited by
-
High genetic diversity and different type VI secretion systems in Enterobacter species revealed by comparative genomics analysis
BMC Microbiology (2024)
-
Synthetic microbe-to-plant communication channels
Nature Communications (2024)
-
The role of the type VI secretion system in the stress resistance of plant-associated bacteria
Stress Biology (2024)
-
Soil bacterium manipulates antifungal weapons by sensing intracellular type IVA secretion system effectors of a competitor
The ISME Journal (2023)
-
Collective protection against the type VI secretion system in bacteria
The ISME Journal (2023)