Abstract
In many ecological communities, predation has a key role in regulating community structure or function. Although predation has been extensively explored in animals and microbial eukaryotes, predation by bacteria is less well understood. Here we show that predatory bacteria of the genus Halobacteriovorax are prevalent and active predators on the surface of several genera of reef-building corals. Across a library of 198 16S rRNA samples spanning three coral genera, 79% were positive for carriage of Halobacteriovorax. Cultured Halobacteriovorax from Porites asteroides corals tested positive for predation on the putative coral pathogens Vibrio corallyticus and Vibrio harveyii. Co-occurrence network analysis showed that Halobacteriovorax’s interactions with other bacteria are influenced by temperature and inorganic nutrient concentration, and further suggested that this bacterial predator’s abundance may be driven by prey availability. Thus, animal microbiomes can harbor active bacterial predators, which may regulate microbiome structure and protect the host by consuming potential pathogens.
Similar content being viewed by others
Introduction
Host–microbe relationships are so important for the health of many plants and animals that the term ‘holobiont’ has been coined to describe the sum of a host and its symbionts (Hosokawa et al., 2006). In addition to their interactions with an animal host, host-associated microbes simultaneously compete and cooperate with one another, and alterations to these interactions can affect holobiont function (De Boer et al., 2007). Although top-down control via predation is known to structure many animal communities (Baum and Worm, 2009), the role of predation by bacteria on the structure of microbial communities is less well studied. Both specialist and generalist predatory strategies impact ecological communities by shifting community composition and abundance, but through different mechanisms. Specialists' predation via viral infections drives an evolutionary arms race within their specific prey population and can destroy entire clonal blooms of phytoplankton (Martínez et al., 2007). Conversely, generalist predators have more influence over the diversity and dynamics of their aggregate prey community.
Bdellovibrionales and like organisms (BALOs) are an order of predatory δ-proteobacteria that prey exclusively on other bacteria, including many known pathogens (Schoeffield and Williams, 1990). Predation by BALOs also releases nutrients (Martínez et al., 2013), affecting biogeochemical cycling and production in nutrient-limited environments, similar to the effects of phage (Fuhrman, 1999; Brussaard et al., 2008). In contrast to viruses, which are the most abundant biological entity in the marine environment and maintain abundances approximately in an order of magnitude greater than their hosts, BALOs do not need to be in high initial concentrations in the environment to drive significant bacterial mortality of their prey (Richards et al., 2012; Williams et al., 2015). One group of Bdellovibrionales, the marine Halobacteriovorax, are broadly distributed in marine waters across temperature, salinity and pH gradients (Pineiro et al., 2007). Halobacteriovorax predation represents a regulatory mechanism that can alter the structure of bacterial communities (Li et al., 2014).
Scleractinian or hard corals are widely studied because of their fundamental role as ecosystem engineers that build tropical reefs. Previous metagenomic and 16S amplicon studies suggest that Halobacteriovorax are members of coral microbiomes, as they were detected in Porites (Wegley et al., 2007) and Acropora species (Sweet and Bythell, 2015). However, active predation by coral-associated Halobacteriovorax strains and the ecological interactions of this genus with other coral-associated microbes remain unexplored. Here we use a combination of cultivation, predation assays, microscopy, and 16S rRNA gene amplicon 454 pyrosequencing (Supplementary Methods) to assess Halobacteriovorax’s role in the microbiomes of three corals (Siderastrea siderea, Agaricia spp. and Porites spp.) in a 3-year data set.
Results and Discussion
We confirmed that Halobacteriovorax are members of coral microbiomes by culturing isolates from the three coral genera sampled in the time-series experiment and Montastraea cavernosa corals adjacent to the experimental plots (Figures 1a–d; Supplementary Table S1). Using full-length 16S analysis, the phylogenetic relatedness of these isolates to other BALOs was inferred (Figure 1e). Our Halobacteriovorax isolates were all identical at the 16S rRNA sequence level (accession #KR493097), and grouped within the previously established Halobacteriovorax cluster XIII (Figure 1e; Pineiro et al., 2007).
Electron micrographs of our coral-associated strain, Halobacteriovorax PA1, including (a) several attack-phase cells and a Vibrio fortis prey (the single larger cell), (b) a single attack-phase cell, (c) a cell in attachment phase on the surface of a Vibrio fortis prey cell and (d) a Halobacteriovorax entering the prey periplasm. (e) Molecular phylogenetic analysis is based on 16S rRNA gene sequences of the order Bdellovibrionales by the maximum likelihood method. The coral-isolated strain (PA1) used in this study is blue, and strains with whole-genome sequences are red. The bootstrap consensus tree inferred from 500 replicates and bootstrap values >80% are reported. The analysis involved 23 nucleotide sequences. All positions containing gaps and missing data were eliminated. There were a total of 1216 positions in the final data set. Evolutionary analyses were conducted in MEGA6 [3].
We then explored whether our strains targeted other members of the coral microbiome for predation by selecting one of our Porites astreoides isolates, Halobacteriovorax sp. PA1, for the plaque assay technique that determines prey range (Schoeffield and Williams, 1990). Halobacteriovorax PA1 successfully attacked and consumed the known coral pathogens Vibrio coralliilyticus and Vibrio harveyi, as well as a Vibrio fortis strain (accession no. AJ440005, CP009468.1 and KT626460, respectively), we isolated from an apparently healthy P. asteroides colony. Like free-living isolates, host-associated Halobacteriovorax followed a biphasic lifecycle consisting of growth and attack phases (Supplementary Figure S1) visible in scanning electron microscopy (SEMs; Figures 1b–d).
We then examined the prevalence of Halobacteriovorax across coral microbiomes using a 3-year time series of 16S rRNA libraries generated from the surface mucus layer of corals (Supplementary Methods) that were exposed to increased nitrogen and phosphorus to simulate nutrient pollution, environmental factors that indirectly contribute to coral disease (Vega Thurber et al., 2014). Halobacteriovorax was prevalent in >79% of all samples (n=198) and not detected in the other samples. Prevalence of Halobacteriovorax in nutrient-enriched corals (81%; n=115) was not significantly different (P=0.28) from controls (78%; n=83); Halobacteriovorax relative abundance was also not significantly different (P=0.14) between the two data sets. The mean relative abundance of Halobacteriovorax across all libraries was low (0.40%±0.04 SEM), similar to their abundances in estuarine systems (Pineiro et al., 2013) and as characteristic of predators across most ecosystems (Baum and Worm, 2009). The high prevalence of this taxon and its predatory lifestyle suggests it is an important low-abundance member of the core coral microbiome (core members are detected in ⩾75% and are consistent components of the microbial assemblage). Given their low abundance, false negatives cannot be established with certainty, and the reported prevalence values likely represent a lower bound on Halobacteriovorax prevalence.
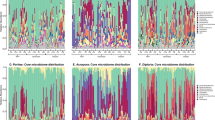
To study the possible interactions between predatory bacteria and other members of the host microbiome, we constructed co-occurrence networks from our coral microbiome data set using the CoNet plugin (Faust and Raes, 2012; Faust et al., 2012) for Cytoscape (Shannon et al., 2003). Networks were generated from the taxonomic order level and provide the net effect of all BALO predation. CoNet uses a permutation-renormalization and bootstrap (ReBoot) method to mitigate the effects of spurious correlations induced by compositionality while assessing the significance of an association. Networks were constructed separately for two known drivers of shifts in the coral microbiome, temperature and nutrient enrichment. Thus, networks were binned by (a) temperature (2 °C intervals) and (b) nutrient enrichment, as well as by (c) coral host (Supplementary Table S2).
Changing thermal regimes affected bacterial predator–prey interactions (Figure 2, Supplementary Table S3). The largest number of significant taxon co-occurrences (network edges) between BALOs and other bacterial taxa occurred at the highest and lowest extremes of temperature (Figure 2). Networks from the highest-temperature data sets formed two distinct clusters. BALOs are a component of the larger cluster in both the control and nutrient networks, where their edges in the network interact with several taxa, including Myxococcales, another predatory bacterial taxa, suggesting that bacterial predators in the network respond to common drivers. Bacterial predator–prey interactions in the networks also fundamentally changed under nutrient enrichment (Figure 2). Only three shared co-occurrence taxa (Chromatiales, NB1-j and unclassfied79) were detected in the networks from the nutrient treatment samples (Supplementary Table S3), and only a single taxa (Cytophagales) was shared between the nutrient and control host network.
Host-associated microbiome networks focused on members of the Bdellovibrionales and like organisms, which include the Halobacteriovorax. Networks were constructed for control samples falling within the temperature bin (a) 23–24 °C and (b) 29–30 °C, and nutrient enrichment samples within the temperature bin (c) 23–24 °C and (d) 29–30 °C. Circles in the networks represent bacterial nodes at order taxa level, color-coded by phylum according to the legend in the center. Blue lines connecting node pairs indicate significant co-occurrence patterns, whereas gray lines denote significant mutual exclusion patterns (P⩽0.05) as determined by the ReBoot randomization procedure in the CoNet Cytoscape package (Faust and Raes, 2012; Faust et al., 2012). Network layout was calculated using edge-weighted spring embedded layout in Cytoscape (Shannon et al., 2003).
Siderastrea and Agaricia coral species contributed a majority of the BALO interactions in the host networks, whereas Porites accounted for only 8% (Supplementary Table S4). Some of the interactions were different among the different coral host. For example, the Bdellovibrionales co-occurred with Vibrionales on Agaricia corals, but not the two other host taxa (Supplementary Table S4). Interestingly, in all the networks generated, BALOs had exclusively positive co-occurrence interactions (blue) and never exhibited mutual exclusions (gray; Figure 2). Co-occurrence interactions suggest that (a) Bdellovibrionales populations are increasing because of the increase in their bacterial prey populations, (b) conditions are favorable to Bdellovibrionales and the co-occurring taxa and/or (c) the predatory bacteria are removing competitors of co-occurring taxa allowing other members to increase. Given the well-known role of prey abundance in other predator–prey systems, response to prey abundance would parsimoniously explain the co-occurrence data. An alternative hypothesis is that low read depth precludes detection of negative interactions by Halobacteriovorax. We examined whether negative interactions were observed for other low-abundance microorganisms. Bacterial relative abundance showed little correlation with the number of negative edges detected (Pearson correlation, r2=0.00159). The same relationship was held when restricting regression analysis to taxa equally or less abundant than Halobacteriovorax (Pearson correlation, r2=0.007).
Active predation by host-associated bacteria opens an intriguing area of research into the structure and function of animal microbiomes. Longitudinal analysis of the coral co-occurrence networks simultaneously delivers both a broad census of the microbiome along with a condensed list of potentially interesting biological interactions that can be targeted in future studies using truly quantitative methods (for example, qPCR of predator–prey cycles). Such truly quantitative approaches would be costly and potentially infeasible without a priori knowledge of which members to target.
Our findings establish that regardless of their low abundance, Halobacteriovorax predation may significantly impact microbiome dynamics in a context-dependent manner and, as a corollary, global change could alter predator–prey dynamics on host-associated microbiomes.
References
Baum JK, Worm B . (2009). Cascading top-down effects of changing oceanic predator abundances. J Anim Ecol 78: 699–714.
Brussaard CPD, Wilhelm SW, Thingstad F, Weinbauer MG, Bratbak G, Heldal M et al. (2008). Global-scale processes with a nanoscale drive: the role of marine viruses. ISME J 2: 575–578.
De Boer W, Wagenaar A-M, Klein Gunnewiek PJA, van Veen JA . (2007). In vitro suppression of fungi caused by combinations of apparently non-antagonistic soil bacteria. FEMS Microbiol Ecol 59: 177–185.
Faust K, Raes J . (2012). Microbial interactions: from networks to models. Nat Rev Microbiol 10: 538–550.
Faust K, Sathirapongsasuti JF, Izard J, Segata N, Gevers D, Raes J et al. (2012). Microbial co-occurrence relationships in the human microbiome. PLoS Comput Biol 8: e1002606.
Fuhrman JA . (1999). Marine viruses and their biogeochemical and ecological effects. Nature 399: 541–548.
Hosokawa T, Kikuchi Y, Nikoh N, Shimada M, Fukatsu T . (2006). Strict host-symbiont cospeciation and reductive genome evolution in insect gut bacteria. PLoS Biol 4: e337.
Li H, Chen C, Sun Q, Liu R, Cai J . (2014). Bdellovibrio and like organisms enhanced growth and survival of Penaeus monodon and altered bacterial community structures in its rearing water. Appl Environ Microbiol 80: 6346–6354.
Martínez JM, Schroeder DC, Larsen A, Bratbak G, Wilson WH . (2007). Molecular dynamics of Emiliania huxleyi and cooccurring viruses during two separate mesocosm studies. Appl Environ Microbiol 73: 554–562.
Martínez V, Jurkevitch E, García JL, Prieto MA . (2013). Reward for bdellovibrio bacteriovorus for preying on a polyhydroxyalkanoate producer. Environ Microbiol 15: 1204–1215.
Pineiro S, Chauhan A, Berhane T-K, Athar R, Zheng G, Wang C et al. (2013). Niche partition of bacteriovorax operational taxonomic units along salinity and temporal gradients in the chesapeake bay reveals distinct estuarine strains. Microb Ecol 65: 652–660.
Pineiro SA, Stine OC, Chauhan A, Steyert SR, Smith R, Williams HN . (2007). Global survey of diversity among environmental saltwater Bacteriovoracaceae. Environ Microbiol 9: 2441–2450.
Richards GP, Fay JP, Dickens KA, Parent MA, Soroka DS, Boyd EF . (2012). Predatory bacteria as natural modulators of Vibrio parahaemolyticus and Vibrio vulnificus in seawater and oysters. Appl Environ Microbiol 78: 7455–7466.
Schoeffield AJ, Williams HN . (1990). Efficiencies of recovery of bdellovibrios from brackish- water environments by using various bacterial species as prey. Appl Envir Microbiol 56: 230–236.
Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D et al. (2003). Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13: 2498–2504.
Sweet M, Bythell J . (2015). White syndrome in Acropora muricata: nonspecific bacterial infection and ciliate histophagy. Mol Ecol 24: 1150–1159.
Vega Thurber RL, Burkepile DE, Fuchs C, Shantz AA, McMinds R, Zaneveld JR . (2014). Chronic nutrient enrichment increases prevalence and severity of coral disease and bleaching. Glob Chang Biol 20: 544–554.
Wegley L, Edwards R, Rodriguez-Brito B, Liu H, Rohwer F . (2007). Metagenomic analysis of the microbial community associated with the coral Porites astreoides. Environ Microbiol 9: 2707–2719.
Williams HN, Lymperopoulou DS, Athar R, Chauhan A, Dickerson TL, Chen H et al. Halobacteriovorax, an underestimated predator on bacteria: potential impact relative to viruses on bacterial mortality. ISME J e-pub ahead of print 7 August 2015; doi:10.1038/ismej.2015.129.
Acknowledgements
We thank numerous volunteers for assistance with the experiment and the Florida Keys National Marine Sanctuary for permits (FKNMS-2009-047 and FKNMS-2011-090). This research was supported by National Science Foundation grant OCE-1130786 to DEB and RVT, Florida International University, and Oregon State University.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on The ISME Journal website
Supplementary information
Rights and permissions
About this article
Cite this article
Welsh, R., Zaneveld, J., Rosales, S. et al. Bacterial predation in a marine host-associated microbiome. ISME J 10, 1540–1544 (2016). https://doi.org/10.1038/ismej.2015.219
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ismej.2015.219
This article is cited by
-
Variance of coral anti-pathogen defense in response to transplantation between coral- and macroalgal-dominated reefs
Coral Reefs (2022)
-
Community and single cell analyses reveal complex predatory interactions between bacteria in high diversity systems
Nature Communications (2021)
-
Host-specific symbioses and the microbial prey of a pelagic tunicate (Pyrosoma atlanticum)
ISME Communications (2021)
-
Identification and analysis of the biological activity of the new strain of Pseudoalteromonas piscicida isolated from the hemal fluid of the bivalve Modiolus kurilensis (F. R. Bernard, 1983)
Archives of Microbiology (2021)
-
Bradymonabacteria, a novel bacterial predator group with versatile survival strategies in saline environments
Microbiome (2020)