Abstract
Accumulating evidence supports that the intestinal microbiome is involved in Type 1 diabetes (T1D) pathogenesis through the gut-pancreas nexus. Our aim was to determine whether the intestinal microbiota in the non-obese diabetic (NOD) mouse model played a role in T1D through the gut. To examine the effect of the intestinal microbiota on T1D onset, we manipulated gut microbes by: (1) the fecal transplantation between non-obese diabetic (NOD) and resistant (NOR) mice and (2) the oral antibiotic and probiotic treatment of NOD mice. We monitored diabetes onset, quantified CD4+T cells in the Peyer’s patches, profiled the microbiome and measured fecal short-chain fatty acids (SCFA). The gut microbiota from NOD mice harbored more pathobionts and fewer beneficial microbes in comparison with NOR mice. Fecal transplantation of NOD microbes induced insulitis in NOR hosts suggesting that the NOD microbiome is diabetogenic. Moreover, antibiotic exposure accelerated diabetes onset in NOD mice accompanied by increased T-helper type 1 (Th1) and reduced Th17 cells in the intestinal lymphoid tissues. The diabetogenic microbiome was characterized by a metagenome altered in several metabolic gene clusters. Furthermore, diabetes susceptibility correlated with reduced fecal SCFAs. In an attempt to correct the diabetogenic microbiome, we administered VLS#3 probiotics to NOD mice but found that VSL#3 colonized the intestine poorly and did not delay diabetes. We conclude that NOD mice harbor gut microbes that induce diabetes and that their diabetogenic microbiome can be amplified early in life through antibiotic exposure. Protective microbes like VSL#3 are insufficient to overcome the effects of a diabetogenic microbiome.
Similar content being viewed by others
Introduction
The initiation and development of Type 1 diabetes (T1D) is not fully understood. In recent years, the incidence rate of T1D increased annually by 3–5% in Western countries (Knip, 2012), suggesting that environmental factors have a role in T1D pathogenesis (reviewed in (Akerblom and Knip, 1998)). The increasing incidence in T1D is most marked in children aged 1–5 years (Vehik et al., 2007), suggesting that early life exposures are critical in shaping the autoimmune response. The ‘hygiene hypothesis’ proposes that insufficient microbial exposure during infancy drives autoimmunity, and certain intestinal microbes and their antigens are hypothesized to induce autoimmune responses. Increased exposure to intestinal microbial antigens or metabolites through the gut could be involved in triggering diabetogenic immune responses (reviewed in (de Kort et al., 2011)).
There is growing appreciation that the intestinal microbiota impacts both health and disease, and compelling associative evidence suggests that gut microbes are involved in the pathogenesis of T1D (Alam et al., 2011; King and Sarvetnick, 2011; Kriegel et al., 2011; Markle et al., 2013; Wen et al., 2008; Yurkovetskiy et al., 2013; Kostic et al., 2015). Infants who develop T1D have a pre-diabetic intestinal microbiota characterized by less diversity and stability, fewer Bifidobacteria spp. and elevated levels of Bacteroides spp. compared with genetically susceptible infants who do not develop T1D (Giongo et al., 2011). Similarly, decreased microbial diversity with fewer Lactobacillus and Bifidobacteria spp., and enriched populations of unclassified Bacteroides spp. have been linked to diabetes development in the Biobreeding diabetes prone rat (Roesch et al., 2009). Segmented filamentous bacteria (SFB) have also been hypothesized to protect non-obese diabetic (NOD) mice against T1D by inducing protective T-helper type 17 (Th17) cell populations in the small intestine (Kriegel et al., 2011). Despite these intriguing findings, our understanding of the potentially causal relationship between gut microbes and T1D is limited. At least two studies show that antibiotics protect against diabetes in disease prone animals: sulfatrim attenuates virus-induced T1D in rats (Hara et al., 2012) and vancomycin treatment attenuates diabetes in NOD mice (Hansen et al., 2012). Although neither of these studies examined the role of the gut barrier in facilitating T1D progression, studies have implicated a leaky gut in the pathogenesis of T1D in both patients and animal models (Sapone et al., 2006; Lee et al., 2010).
In the current study, we hypothesized that the intestinal microbiota promotes pancreatic inflammation and drives the destruction of β-cells. We found the NOD mouse gut microbiota harbored more pathobionts and fewer beneficial microbes in comparison with the diabetes-resistant non-obese resistant (NOR) mouse. Moreover, NOD fecal microbiota when transplanted into NOR mice promoted insulitis. Furthermore, life-long antibiotic exposure from birth accelerated diabetes in NOD mice through altered effector T-cell populations in the gut lymphoid tissues. The antibiotic-induced diabetes correlated with changes to the microbiome characterized by reduced diversity, enriched pathobionts, depletion of protective microbes and overall changes to the metagenome, including several gene clusters involved in metabolism. Similarly, we found that antibiotic treatment resulted in reduced microbial short-chain fatty acid (SCFA) metabolites, which are known to protect against diabetes (Al-Lahham et al., 2010; Mitrou et al., 2015). Finally, to try and overcome the diabetogenic effects of the microbiome, we administered VSL#3 probiotics to NOD mice and found they had no effect on diabetes onset due to their poor colonization of the gut. Taken together, our data reveals that the NOD microbiome is diabetogenic and can be potentiated through antibiotics but not overcome with probiotic treatment. Overall, these results highlight the complexity of the role played by the intestinal microbiota in the modulation of autoimmune diabetes onset in NOD mice.
Materials and methods
Mice
NOD and NOR mice (Jackson Laboratory, Bar Harbor, ME, USA) were housed and bred under specific pathogen-free conditions at the Child and Family Research Institute (Vancouver, BC, Canada). Mice were maintained in a temperature controlled (22±2 °C) animal facility with a 12 h light/dark cycle. Animals received a sterile chow diet (Laboratory Rodent Diet 5001, Purina Mills, St Louis, MO, USA) and filtered UV sterilized water ad libitum throughout experiments. All procedures were approved by the UBC Animal Care Committee under the direct guidelines of the Canadian Council on the Use of Laboratory Animals.
Oral fecal bacterial transfer
Similar to the approach of Ghosh et al. (2011), for fecal transplantation experiments, NOD and NOR mice were treated with antibiotics (500 mg/l vancomycin, 1 g l−1 neomycin, 1 g l−1 ampicillin and 1 g l−1 metronidazole) in drinking water from 4–8 weeks of age. Stool from donor mice (100 mg in 1.2 ml phosphate-buffered saline) was orally gavaged into recipient mice four times over 12 days starting at 8 weeks of age, creating chimeric mice (Figure 2a).
Antibiotic and VSL#3 treatment
Vancomycin (500 mg l−1, Amresco, Soloh, OH, USA), neomycin (1 g l−1, Gold Biotechnology, St Louis, MO, USA) orVSL#3 (1.25 g l−1, VSL#3 Pharmaceuticals, Rome, Italy) containing Streptococcus thermophilus DSM24731, Bifidobacterium breve DSM 24732, B. longum DSM 24736, B. infantis DMS 24737, L. acidophilus DSM 24735, L. plantarum DSM 24730, L. paracasei DSM 24733 and L. delbrueckii subsp. b ulgaricus DSM 24734 were prepared according to the manufacturer’s instructions and administered in the drinking water of pregnant mice just prior to birth, throughout lactation and continued throughout the lifetime of the pups until diabetes onset. Mice were monitored for diabetes with weekly tail vein blood glucose measurements and euthanized following two consecutive daily readings of >14 mmol l−1.
Tissue, sera and stool collection
Mice were anaesthetized using isofluorane, blood was collected via cardiac puncture and serum was separated by centrifugation. Mice were killed by cervical dislocation, and the terminal ileum, distal colon and pancreas were removed. For T-cell analysis, the Peyer’s patch lymph nodes were collected from pre-diabetic mice that were 4–6 weeks of age, homogenized, and cells filtered through 50 μm cell strainers followed by a phosphate-buffered saline wash. For histology, gut and pancreas tissues from 11–20-week-old mice were immersed in 10% neutral-buffered formalin and processed for scoring and immunofluorescence. For microbial analysis, colon and ileum segments were removed from 11–20-week-old mice, immersed in LN2 and stored at −80 °C. For SCFA analysis, feces was collected from mice that were 10 weeks of age, immersed in LN2 and stored at −80 °C.
T-cell analysis
Similar to Zhang et al. (2010), Peyer’s patch lymphocytes were cultured in a 5 ml tube containing 1 ml of 10% fetal bovine serum or RPMI medium with 50 ng phorbol 12-myristate 13-acetate, 1 μg ionomycin and Golgistop for 4–5 h. Fixation/permeabilization solution (eBioscience, San Diego, CA, USA) was then added overnight. After washing with permeabilization solution, the cells were labeled with fluorescence-conjugated monoclonal antibodies (BD Pharmingen, San Jose, CA, USA): anti-IFNγ-APC, anti-IL17-PE and anti-CD4-Pacific Blue. Fluorescence-activated cell sorting (LSR Fortessa (BD Bioscience, San Jose, CA, USA)) was performed using Flowjo software (Ashland, OR, USA).
Insulitis scoring
Paraffin-embedded sections of pancreas were stained with haematoxylin and eosin and examined by light microscopy (Zeiss Imager. Z1, AXIO, Oberkochen, Germany). All islets were counted and the percentage of islets that had no insulitis, peri-insulitis, invasive insulitis (<50% islet) and invasive insulitis (>50% islet), score 0, 1, 2 and 3, respectively, were determined by two investigators blinded to treatment groups.
Insulin auto-antibody assay
Insulin auto-antibodies were quantified in the sera by the Barbara Davis Center (Aurora, CO, USA).
High throughput sequencing
DNA was extracted from tissue segments of the colon and ileum using a Stool DNA extraction kit (Qiagen, Toronto, ON, Canada). Bacterial DNA from 11- to 20-week-old mice was amplified using universal bacterial primers that flank the V3 and V4 regions of the 16S rRNA gene (Herlemann et al., 2011) with sample-specific barcode sequences (Supplementary Table S1). Following PCR, samples were gel extracted and the concentration normalized to 1.25 ng DNA per μl using Sequalprep (Invitrogen, Burlington, ON, Canada). Sequencing was performed at LAGA (Vancouver, BC, Canada) using Roche 454 FLX+ technology. Sequences were analyzed using the QIIME (Knight lab, Boulder, CO, USA) pipeline (Caporaso et al., 2010). 406 120 sequences (200–600 bp in length and minimum quality score of 25) were denoised and chimera detection (usearch61 (Edgar, 2010)) resulted in the removal of 42 066 sequences. Operational taxonomic units (OTU) were picked (usearch61, 97% similarity threshold) and a representative set of sequences was aligned (MUSCLE (Edgar, 2004)). Taxonomy was assigned using ribosomal database project with the Greengenes database (97% similarity OTUs with the sequence for Candidatus arthromitus (SFB) added). OTU tables were rarefied to 105 sequences. α-diversity was determined using the Chao1 statistic (Chao et al., 2005) and log transformed with zero values linearly adjusted followed by Bray–Curtis (Bray and Curtis, 1957) for β-diversity. Significance was determined using permutational analysis of variance (Anderson, 2001) based on the maximum number of permutations to 9999.
Metagenomic analysis
Metagenome function of the 16S libraries were predicted using PICRUSt (Phylogenetic Investigation of Communities by Reconstruction of Unobserved States, version 1.0.0; Langille et al., 2013). Closed reference OTUs were picked against Greengenes database (May 2013) with 97% percent similarity. OTUs were normalized for copy number and the gene families predicted at level 2 and level 3 KEGG orthology groups (Kanehisa et al., 2012). The data were normalized to 27 500 genes per sample and similarity in the gene families between samples determined by Bray–Curtis similarity on log-transformed data. Differences between groups are determined by permutational analysis of variance using the maximum number of permutations to 9999.
SCFA analysis
The amount of acetic, propionic and butyric acid was analyzed from fecal samples collected from pre-diabetic mice by direct-injection gas chromatography. Fecal samples were homogenized in isopropyl alcohol, containing 2-ethylbutyric acid at 0.01% v/v used as an internal standard and then centrifuged. This step was repeated. The supernatant was injected into a Trace 1300 Gas Chromatograph, equipped with flame-ionization detector, with AI1310 autosampler (Thermo Scientific, Walkham, MA, USA) in splitless mode. A fused-silica FAMEWAX (Restekas, Bellefonte, PA, USA) column 30 m × 0.32 mm i.d. coated with 0.25 μm film thickness was used. Helium was supplied as the carrier gas at a flow rate of 1.8 ml min−1. The initial oven temperature was 80 °C, maintained for 5 min, raised to 90 °C at 5 °C min−1, then increased to 105 °C at 0.9 °C min−1, and finally increased to 240 °C at 20 °C min−1 and held for 5 min. The temperature of the flame-ionization detector and the injection port was 240 °C and 230 °C, respectively. The flow rates of hydrogen, air and nitrogen as makeup gas were 30, 300 and 20 ml min−1, respectively. Data were analyzed with Chromeleon 7 software (Bannockburn, IL, USA). Fine separation of SCFA was confirmed by the complete separation of the volatile-free acid mix (Sigma, Oakville, ON, Canada). Data are presented as mass % (g of SCFA per g of feces × 100).
Statistical analysis
Graphing and statistical analysis was performed using Graphpad Prism (La Jolla, CA, USA), QIIME and PRIMER-E (Lutton, UK) software. For comparisons between two groups, *P<0.05, **P<0.01 and ***P<0.0001 by unpaired t-test for parametric data or Mann–Whitney's U-test for non-parametric data. For comparisons between >2 groups, *P<0.05, **P<0.01 and ***P<0.0001 by one-way analysis of variance with Tukey’s post hoc test for parametric data and Kruskal–Wallis test with Dunn’s post hoc test for non-parametric data. Where multiple comparisons were drawn from the same data same data for metagenomic sequencing analysis, false discovery rate-corrected analysis of variance with Tukey’s post hoc test was used. Mantel-cox was used for survival analysis and permutational analysis of variance was used for microbial community analysis.
Results
Diabetes-susceptible and diabetes-resistance mice harbor distinct intestinal microbiota
The intestinal microbiota from the colon and ileum of diabetes susceptible (NOD) and diabetes resistant (NOR) mice were profiled via 454 pyrosequencing. NOR and NOD mice harbor distinct microbial communities in the ileum and colon (Figures 1a and b). Although the microbiota was similar at the phyla level (1c), several differences were detected at lower taxonomic levels (Figure 1d and e). In the ileum, the NOD microbiota was comparatively reduced in SFB and Lactobacillus spp., while Anaeroplasma spp. and Desulfovibrio spp. were present in the ileums of NOD mice and absent in NOR (Figure 1d). In the colon, NOD mice had fewer members of the Alphaproteobacteria class, no Bacteroides acidifaciens and fewer Ruminococcus gnavus compared with NOR mice. Prevotella spp. were present in the colon of NOD mice and not detected in NOR (Figure 1e). In general, the NOR microbiota had more microbes previously described as beneficial to the host, whereas the NOD microbiota had more pathobionts, microbes that have pathogenic qualities.
NOD and NOR mice harbor different gut microbiota. Microbial communities from the colon and ileum were assessed by 454 pyrosequencing. (a and b) The microbiota from NOR and NOD mice differed from each other both in the ileum (a; P=0.03) and the colon (b; P=0.009). (c) At the phyla level, NOR and NOD microbiota is similar yet the abundance of certain taxa is different in NOD versus NOR in the ileum (d) and colon (e; n=5-8; ND=none detected). *P<0.05 by FDR-corrected one way analysis of variance with Tukey's post hoc test.
The NOD mouse harbors microbiota that can exacerbate insulitis in NOR mice
To determine whether the intestinal microbiota of the NOD mouse is a causal factor in its development of T1D, we performed a series of fecal oral transplants using NOD and NOR mice. We generated four groups of chimeric mice: NORH+NORM, NORH+NODM, NODH+NODM and NODH+NORM (H, host; M, microbiota; Figure 2a) by treating hosts with a combination of antibiotics followed by oral gavage of stool from donor mice as previously described (Ghosh et al., 2011). Normally, NOR mice develop limited insulitis (Prochazka et al., 1992), however, when transplanted with microbiota from NOD mice, the NORH+NODM mice displayed greater insulitis compared with control NORH+NORM mice (Figures 2b–d). In contrast, the microbiome from the NOR host were unable to significantly protect against insulitis when transplanted into the NOD mouse, as evidenced by similar levels of insulitis between NODH+NORM and NODH+NODM (Figures 2b–d). We examined the abundance of microbes in the intestine before and after transplantation confirming the fecal transplantation of the microbiota in chimeric mice (Supplementary Figure 1). These results suggest the NOD mice harbor a microbiome capable of inducing pancreatic inflammation in normally resistant mice. However, the microbiome from NOR mice cannot outcompete the diabetogenic microbes to overcome the genetic predisposition to insulitis in NOD mice.
Mice susceptible to diabetes harbor a microbiota that initiates insulitis in diabetes-resistant mice. (a) Schematic representation of fecal transplantation between NOR and NOD mice. Tissues were collected at 11 weeks and cross-sections of pancreas tissues were stained with H&E and scored by two blinded observers. (b) Representative pictures of islet infiltration. (c) Composite insulitis score and (d) percentage of islets in each mouse that were scored 0, 1, 2 and 3 (scale bar=100 μm; n=10–12 mice per group). *P<0.05 by Mann-Whitney U-test.
Antibiotic exposure accelerates diabetes in NOD mice
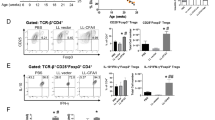
As the NOD mouse microbiome caused exaggerated insulitis, we wanted to determine whether we could eliminate the diabetogenic microbes through antibiotic treatment and thereby protect against diabetes onset. To examine this, we gave pregnant NOD mice vancomycin or neomycin via the oral–gastric route just prior to them giving birth, and continued this treatment throughout weaning and then to the offspring until diabetes onset. Despite our hypothesis, we found that both vancomycin and neomycin accelerated diabetes in NOD mice when compared with NOD mice given water only (Figure 3a). Similar to our previous studies showing ~30% of NOD mice are diabetic by 20 weeks (Lee et al., 2011), ∼70% of antibiotic treated NOD mice were diabetic by 20 weeks. In support of this accelerated diabetes, the antibiotic-treated mice had more serum insulin auto-antibodies, and altered gut lymphoid-associated effector T cells including increased interferon-γ+ CD4+ T cells and reduced interleukin-17+ CD4+ T cells (Figures 3b–d). These results suggest that the establishment of the intestinal microbiota early in life is important in controlling the diabetogenic microbiome and effector T-cell populations in the gut-lymphoid tissues.
Antibiotic exposure accelerates diabetes in NOD mice and alters CD4+ T-cell populations. (a) Vancomycin and neomycin accelerated diabetes onset in NOD mice (n=17–25) *P<0.05 and **P<0.01 by log-rank survival test. (b) Serum insulin auto-antibody levels in pre-diabetic 11-week old NOD mice (n=10). (c) Percent of IFN-γ+ CD4+ T cells and (d) percent of IL17+ CD4+ T cells in the Peyer’s patches of 4–6 week old NOD mice (n=12). *P<0.05, **P<0.01 and ***P<0.001 by one way analysis of variance with Tukey's post hoc test for parametric data and Kruskal-Wallis test with Dunn's post hoc test for non parametric data.
Antibiotic-induced diabetes is associated with microbial dysbiosis
To determine which microbes were associated with antibiotic-accelerated diabetes, the microbial populations of antibiotic-treated mice were profiled by 454 pyrosequencing. Following antibiotic treatment, we found that richness of the microbes decreased (Figure 4a) and that vancomycin and neomycin treatments resulted in distinct microbial communities in the ileum (Figure 4b) and colon (Figure 4c). Several microbes emerged as key players in driving the differential clustering between groups in the ileum and colon (Figures 4d and e and Supplementary Figure 2). Both antibiotic treatments resulted in the loss of Desulfovibrio spp. in the ileum and Prevotella spp., B. acidifaciens, R. gnavus and Roseburia spp. in the colon (Figures 4d and e and Supplementary Figure 2). Beyond this, the profiles of the vancomycin- and neomycin-treated mice were different. Vancomycin-treated mice had fewer members of Bacteroides S24-7 in the ileum and colon and fewer Ruminococcus spp., Clostridia spp. and Lachnospiraceae in the colon. Increased abundance of Akkermansia muciniphila and Enterobacteriaceae was observed throughout the gut in the vancomycin-treated mice, while Sutterella and Anaeroplasma spp. were increased in the ileum and colon, respectively. The microbial community of the neomycin-treated mice was depleted in several microbes that were increased with vancomycin treatment; Enterobacteriaceae, Anaeroplasma spp. and Sutterella spp., while an increase in the Rickennellaceae family was observed. These results reveal that both antibiotics accelerate diabetes, and although several microbes are similarly depleted with either antibiotic treatment, there appears to be no specific microbe that emerges as a driving force or initiator of autoimmunity.
Antibiotic-induced diabetes is associated with microbial dysbiosis. Communities were profiled in the ileum and colon of control NOD mice and those treated with vancomycin and neomycin by 454 pyrosequencing. (a) Antibiotic-treated mice have less diverse intestinal microbiota. Microbial communities of antibiotic-treated mice are different compared with untreated NOD in the ileum (b; vancomycin P=0.003, neomycin P=0.003) and colon (c; vancomycin P=0.004, neomycin P=0.005). Several microbes are different in abundance in the ileum (d) and colon (e) of antibiotic-treated mice (n=4–8, ND=none detected). *P<0.05 and **P<0.01 by FDR-corrected one way analysis of variance with Tukey's post hoc test.
The diabetogenic microbiome has altered metabolism
To determine whether a particular set of microbial genes were similarly altered in the diabetogenic microbiome, we examined the predicted gene clusters based on the 16S 454 pyrosequencing data using database genomes. Although the mice treated with neomycin clustered similarly to the NOD microbial metagenome, the vancomycin-treated mice clustered distinctly (Figures 5a and b). Despite the microbial metagenome of the neomycin group clustering similarly to the NOD group, both antibiotic-treated groups showed altered metabolic genes including a reduction in several gene clusters involved in the biosynthesis of SCFA including starch and sugar metabolism (Figures 5c and d). SCFA are the major end products of bacterial metabolism in the large intestine including acetate, propionate and butyrate. To determine whether the diabetogenic microbial metagenome resulted in altered SCFA metabolism, we examined fecal SCFA production in the NOR and NOD mice (Figure 6). Our data reveal that SCFAs are reduced in NOD mice treated with antibiotics. Although vancomycin-treated mice had reduced levels of all the major SCFAs, neomycin similarly reduced levels of only acetic acid. Moreover, acetic acid was highly elevated in the NOR mice when compared with the NOD mice. These results suggest that the diabetogenic microbiome lacks the genes that produce SCFAs like acetic acid, potentially underlying the accelerated diabetes.
Metagenomic analysis of microbiota from antibiotic-treated NOD mice. PCoA plots showing clustering of predicted KEGG orthologs present in the microbiota of untreated, vancomycin-treated and neomycin-treated NOD mice in the ileum (a; neomycin P=0.59, vancomycin P=0.014) and distal colon (b; neomycin P=0.263, vancomycin P=0.001). (c) The microbiome’s metabolic gene groupings in the ileum and distal colon and (d) differentially abundant gene groupings present in the colonic microbiome of NOD mice (n=4–8 per group). *P<0.05 and **P<0.01 by FDR-corrected one way analysis of variance with Tukey's post hoc test.
NOD mice given antibiotics carry a microbiota that produces fewer short-chain fatty acids. The fecal content of NOR mice contains more acetic acid than NOD mice and antibiotic treatment further reduces the acetic acid content. Vancomycin, but not neomycin, treatment reduces the amount of propionic and butyric acids found in the cecal content (n=12 mice/group). *P<0.05 and ***P<0.001 by one way analysis of variance with Tukey's post hoc test for parametric data and Kruskal-Wallis test with Dunn's post hoc test for non parametric data.
Probiotic VSL#3 bacteria cannot overcome the effects of the diabetogenic microbiome
We hypothesized that the lack of beneficial microbes in NOD mice contributed to diabetes onset. VSL#3 contains several beneficial microbes, which were not present in NOD mice but present in NOR mice (Figure 1) including taxa from Lactobacillus, Bifidobacteria and Streptococcus. To determine whether we could augment protective microbes and delay T1D, we exposed neonates to VSL#3 in drinking water initially through their mother and then also post-natally until diabetes onset. We found that VSL#3 did not delay diabetes onset (Figure 7a). To understand whether this was due to in the inability of VSL#3 to colonize the gut, we examined the mouse tissues for the presence of VSL#3 microbes and found that colonization was inefficient (Figure 7b–d). Lactobacillus and Bifidobacteria did not colonize either the ileum or the colon and Streptococcus was found to colonize only the colon but not the ileum. We conclude that VSL#3 cannot overcome the effects of the diabetogenic microbiome due to their inability to efficiently colonize the gut.
VSL#3 does not alter diabetes progression and VSL#3 bacteria do not efficiently colonize efficiently. (a) VSL#3 does not alter the onset of diabetes in NOD mice (n=17–25 mice per group). (b–d) qPCR was performed on gut tissues taken from mice untreated or treated with VSL#3 in drinking water. (b) Lactobacillus and (c) Bifidobacteria spp. were not increased in either the ileum or colon. Streptococcus spp. were not increased in the ileum but were increased in the colon (n=10–12 mice per group). *P<0.05 by Mann-Whitney U-test.
Discussion
Autoimmune destruction of β-cells and the loss of endogenous insulin that occurs during T1D is a result of immune dysfunction within the initiation, propagation and/or regulatory stages that control normal immune responses. Although it is unknown what triggers the dysfunctional immune responses in T1D, innate microbial sensing systems within the intestine are involved in the induction of diabetes (Wen et al., 2008). Furthermore, pancreatic lymph nodes contain T cells that originate in the lymphatic tissues of the gut where they are primed to become auto-reactive (Jaakkola et al., 2003). In this study, we explored the relationship between the intestinal microbiota and the onset of T1D in the NOD mouse. We found that NOD mouse fecal microbiota exacerbated pancreatic inflammation when transplanted into normally diabetes-resistant mice. Moreover perinatal antibiotic exposure led to accelerated diabetes onset revealing that the diabetogenic microbiome is established early in life. This was in contrast to our use of postnatal antibiotic exposure that did not induce accelerated diabetes (data not shown). The diabetogenic microbiome carried a reduced capacity for SCFA metabolism associated with increased Th1 and reduced Th17 cells in the gut lymphoid tissues. In an attempt to correct the diabetogenic microbiome, we administered VLS#3 probiotics to NOD mice but we found that microbes in VSL#3 colonized the intestine poorly and did not delay diabetes.
The predisposition to TID in NOD mice may be facilitated by abnormalities in the gut microbiota. Microbes were first suggested to be involved in the development of T1D when it was demonstrated that a high diabetes incidence in NOD mouse colonies depended on the microbiological cleanliness of the animal facility (Pozzilli et al., 1993). Here we found that NOD mice harbor different microbiota than resistant NOR mice, as characterized by specific microbes in the colon and ileum. The ileum microbiota of NOD mice was characterized by fewer SFB and Lactobacillus spp., both previously found to protect against T1D (Valladares et al., 2010; Kriegel et al., 2011; Lau et al., 2011). In the colon, there were no B. acidifaciens detected in NOD mice and fewer R. gnavus. B. acidifaciens induces production of secretory IgA in the gut (Yanagibashi et al., 2013), while R. gnavus has been shown to make anti-inflammatory bile acid (Lee et al., 2013). Despite the differences in microbial profiles, a causative role for the microbiota in T1D progression in NOD mice has not yet been established. Here we show that transplanting NOD microbiota into resistant NOR mice, results in an increase in insulitis in NOR mice suggesting the NOD mice harbor a diabetogenic ecosystem. However, we could not decipher if the changes in insulitis were due to the microbiota or microbial products.
In the present study, we found that antibiotic exposure amplified diabetes-causing microbial patterns resulting in accelerated diabetes onset. This is similar to a recently published study showing that antibiotics in early life alter the gut microbiome and increase diabetes incidence in NOD male mice (Candon et al., 2015). We also observed that male mice had a trend toward increased diabetes incidence (data not shown). There were, however, some notable differences in the experimental design between the two studies: in our study, the concentration of vancomycin in drinking water was 2.5 × higher, we did not treat mating pairs to avoid the influence of the altered male microbiome and we did not treat pregnant dams throughout gestation to avoid teratogenic effects on the fetus. Indeed, our study and Candon et al. (2015) show differences in the microbiome although Candon et al analyzed the microbiota from feces and our study examined gut tissues. In contrast to both of these studies, Hansen et al., (2012) found that vancomycin treatment attenuated diabetes in the NOD mouse. The difference in outcomes may be attributed to the variations in the protocol where we induced continuous dysbiosis by exposing mice to vancomycin from birth until diabetes onset whereas they exposed mice from birth only until weaning. Despite the protocol and phenotype differences, vancomycin treatment in our study and in Hansen et al., (2012) resulted in similar global changes in microbes. Here, we show that the microbiota following either vancomycin or neomycin treatment was similarly less diverse than untreated controls, but were each associated with a unique microbial profile. Although no causative microbe emerged from our analysis, both antibiotic treatments resulted in the loss of Desulfovibrio spp. in the ileum, as well as loss of Prevotella spp., B. acidifaciens, R. gnavus and Roseburia spp. in the colon. Inter-facility variation is often reported in NOD mice, with diabetes incidence ranging from 10 to 90% at 20 weeks and 20 to 100% at 30 weeks of age (Pozzilli et al., 1993). As our colony has a lower incidence of diabetes relative to colonies in some other facilities, the destabilization/loss of protective microbes may also contribute to accelerated diabetes in the present study.
A recent study has shown that a given species is highly variable in infants predisposed to T1D, however, metabolic composition and metabolic pathway redundancy is constant (Kostic et al., 2015). Indeed, we also found the metagenome of diabetogenic microbes were similarly reduced in clusters of genes involved in microbial metabolism. These genetic pathways are involved in the biosynthesis of SCFA production (Macfarlane and Macfarlane, 2003). SCFA are produced by the microbiota and are known to alter metabolism and diabetes outcomes as well as improve intestinal homeostasis and protect against chronic intestinal inflammation (Al-Lahham et al., 2010; Puertollano et al., 2014). In particular, we found that the increased acetic acid correlated with resistance to diabetes. Acetic acid, or vinegar, has been shown to improve insulin resistance and metabolic abnormalities during pre-diabetes in human clinical trials (Mitrou et al., 2015). Although we correlate the lack of SCFA with diabetes onset in this study, further experimental evidence will be required to determine whether acetic acid has a protective effect upon diabetogenesis.
The ‘leaky gut’ theory has been suggested as a potential mechanism by which increased exposure of the immune system to luminal microbial antigens could result in the stimulation of autoimmune responses leading to T1D (de Kort et al., 2011). However, we did not find that antibiotic-induced diabetes was associated with barrier dysfunction (Supplementary data Figure 3). Instead, we hypothesize that in a genetically susceptible host, reduced levels of SCFA deregulate intestinal immune responses that initiate autoimmune response through the gut-pancreas nexus. In a diabetes-resistant host, acetic acid may directly signal through the intestinal barrier as it is known that acetate is available to the host systemically (Pomare et al., 1985). Indeed, SCFA regulate the differentiation of peripheral T cells (Kim et al., 2014). In support of this, we found that the antibiotic treated NOD mice had altered gut lymphoid effector T cells with increased Th1 cells and decreased Th17 cells. Similarly, Candon et al. (2015) also found a lack of Th17 cells was associated with antibiotic-induced diabetes. Based on previous studies showing Th1 cells are pro-diabetic (Rabinovitch et al., 1995), the antibiotic-induced shift toward increased interferon-γ production by CD4+ T cells in the Peyer’s patches could be a key factor in driving insulitis in the pancreas of the antibiotic treated NOD mice. Moreover, as Th17 cells have been shown to be important in protecting against T1D (Kriegel et al., 2011), having decreased interleukin-17+ CD4 T cells suggests that antibiotics also lead to impairment in Th17 dependent protection within the gut. These data reveal that increased pro-inflammatory Th1 cells and a lack of protective Th17 cells from the gut could be important in driving insulitis in the pancreas.
Although little is known about the genesis of Th17 cells in T1D patients, interleukin-17-producing cells in the lamina propria are promoted by SFB in mice (Ivanov et al., 2008, Kriegel et al., 2011). Moreover, SFB have been shown to induce testosterone (Yurkovetskiy et al., 2013), which protects female NOD mice from diabetes (Markle et al., 2013). In our study, we found that vancomycin-treated mice suffered a loss of SFB. However, we did not see a loss of SFB with neomycin treatment suggesting SFB may not be on the only microbe involved in regulating Th17 cells in the gut lymphoid tissues. Furthermore, we did not find any differences in testosterone levels during antibiotic-induced diabetes (data not shown).
It has been hypothesized that beneficial microbes or probiotics can protect against diabetes (Neu et al., 2010). VSL#3 has been shown to prevent diabetes in NOD mice when administered after weaning (Calcinaro et al., 2005). Yet, administration to germ-free NOD neonatal mice shows no effect (Yurkovetskiy et al., 2013). Although NOD mice were deficient in several beneficial microbes present in the probiotic VSL#3, we found that the administration of VSL#3 failed to protect against accelerated diabetes in NOD mice. Despite VSL#3 being administered prior to the establishment of the microbiome, we show that the probiotics colonized the gut inefficiently. Improving the ability of VSL#3 probiotics to colonize the intestine may be important to regulate the beneficial effects protecting against diabetes. Overall, we conclude that VSL#3 cannot overcome the effects of the diabetogenic microbiome due to their inability to efficiently colonize the gut.
The relationship between intestinal microbes and T1D pathogenesis is complex, multifaceted and needs to be studied in more detail. We show that a diabetogenic microbiome facilitates diabetes onset through the gut-pancreas nexus via microbial metabolites. The diabetogenic microbiome can be potentiated through the use of antibiotics highlighting that caution is required for the overuse of antibiotics in infants known to be at risk for T1D. Understanding which microbes induce or conversely protect against T1D could help in the production of more specific antimicrobials as well as effective probiotics.
References
Akerblom HK, Knip M . (1998). Putative environmental factors in Type 1 diabetes. Diabetes Metab Rev 14: 31–67.
Al-Lahham SH, Peppelenbosch MP, Roelofsen H, Vonk RJ, Venema K . (2010). Biological effects of propionic acid in humans; metabolism, potential applications and underlying mechanisms. Biochim Biophys Acta 1801: 1175–1183.
Alam C, Bittoun E, Bhagwat D, Valkonen S, Saari A, Jaakkola U et al. (2011). Effects of a germ-free environment on gut immune regulation and diabetes progression in non-obese diabetic (NOD) mice. Diabetologia 54: 1398–1406.
Anderson MJ . (2001). A new method for non-parametric multivariate analysis of variance. Austral Ecol 26: 32–46.
Bray JR, Curtis JT . (1957). An Ordination of the Upland Forest Communities of Southern Wisconsin. Ecol Monogr 27: 326–349.
Calcinaro F, Dionisi S, Marinaro M, Candeloro P, Bonato V, Marzotti S et al. (2005). Oral probiotic administration induces interleukin-10 production and prevents spontaneous autoimmune diabetes in the non-obese diabetic mouse. Diabetologia 48: 1565–1575.
Candon S, Perez-Arroyo A, Marquet C, Valette F, Foray AP, Pelletier B et al. (2015). Antibiotics in early life alter the gut microbiome and increase disease incidence in a spontaneous mouse model of autoimmune insulin-dependent diabetes. PLoS One 10: e0125448.
Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK et al. (2010). QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7: 335–336.
Chao A, Chazdon RL, Colwell RK, Shen TJ . (2005). A new statistical approach for assessing similarity of species composition with incidence and abundance data. Ecol Lett 8: 148–159.
de Kort S, Keszthelyi D, Masclee AA . (2011). Leaky gut and diabetes mellitus: what is the link? Obes Rev 12: 449–458.
Edgar RC . (2004). MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics 5: 113.
Edgar RC . (2010). Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26: 2460–2461.
Ghosh S, Dai C, Brown K, Rajendiran E, Makarenko S, Baker J et al. (2011). Colonic microbiota alters host susceptibility to infectious colitis by modulating inflammation, redox status, and ion transporter gene expression. Am J Physiol Gastrointest Liver Physiol 301: G39–G49.
Giongo A, Gano KA, Crabb DB, Mukherjee N, Novelo LL, Casella G et al. (2011). Toward defining the autoimmune microbiome for type 1 diabetes. ISME J 5: 82–91.
Hansen CH, Krych L, Nielsen DS, Vogensen FK, Hansen LH, Sorensen SJ et al. (2012). Early life treatment with vancomycin propagates Akkermansia muciniphila and reduces diabetes incidence in the NOD mouse. Diabetologia 55: 2285–2294.
Hara N, Alkanani AK, Ir D, Robertson CE, Wagner BD, Frank DN et al. (2012). Prevention of virus-induced type 1 diabetes with antibiotic therapy. J Immunol 189: 3805–3814.
Herlemann DP, Labrenz M, Jurgens K, Bertilsson S, Waniek JJ, Andersson AF . (2011). Transitions in bacterial communities along the 2000 km salinity gradient of the Baltic Sea. ISME J 5: 1571–1579.
Ivanov II, Frutos Rde L, Manel N, Yoshinaga K, Rifkin DB, Sartor RB et al. (2008). Specific microbiota direct the differentiation of IL-17-producing T-helper cells in the mucosa of the small intestine. Cell Host Microbe 4: 337–349.
Jaakkola I, Jalkanen S, Hanninen A . (2003). Diabetogenic T cells are primed both in pancreatic and gut-associated lymph nodes in NOD mice. Eur J Immunol 33: 3255–3264.
Kanehisa M, Goto S, Sato Y, Furumichi M, Tanabe M . (2012). KEGG for integration and interpretation of large-scale molecular data sets. Nucleic Acids Res 40: D109–D114.
Kim CH, Park J, Kim M . (2014). Gut microbiota-derived short-chain Fatty acids, T cells, and inflammation. Immune Netw 14: 277–288.
King C, Sarvetnick N . (2011). The incidence of type-1 diabetes in NOD mice is modulated by restricted flora not germ-free conditions. PLoS One 6: e17049.
Knip M . (2012). Descriptive epidemiology of type 1 diabetes-is it still in? Diabetologia 55: 1227–1230.
Kostic AD, Gevers D, Siljander H, Vatanen T, Hyotylainen T, Hamalainen AM et al. (2015). The dynamics of the human infant gut microbiome in development and in progression toward Type 1 diabetes. Cell Host Microbe 17: 260–273.
Kriegel MA, Sefik E, Hill JA, Wu HJ, Benoist C, Mathis D . (2011). Naturally transmitted segmented filamentous bacteria segregate with diabetes protection in nonobese diabetic mice. Proc Natl Acad Sci USA 108: 11548–11553.
Langille MG, Zaneveld J, Caporaso JG, McDonald D, Knights D, Reyes JA et al. (2013). Predictive functional profiling of microbial communities using 16 S rRNA marker gene sequences. Nat Biotechnol 31: 814–821.
Lau K, Benitez P, Ardissone A, Wilson TD, Collins EL, Lorca G et al. (2011). Inhibition of type 1 diabetes correlated to a Lactobacillus johnsonii N6.2-mediated Th17 bias. J Immunol 186: 3538–3546.
Lee AS, Gibson DL, Zhang Y, Sham HP, Vallance BA, Dutz JP . (2010). Gut barrier disruption by an enteric bacterial pathogen accelerates insulitis in NOD mice. Diabetologia 53: 741–748.
Lee AS, Ghoreishi M, Cheng WK, Chang TY, Zhang YQ, Dutz JP . (2011). Toll-like receptor 7 stimulation promotes autoimmune diabetes in the NOD mouse. Diabetologia 54: 1407–1416.
Lee JY, Arai H, Nakamura Y, Fukiya S, Wada M, Yokota A . (2013). Contribution of the 7beta-hydroxysteroid dehydrogenase from Ruminococcus gnavus N53 to ursodeoxycholic acid formation in the human colon. J Lipid Res 54: 3062–3069.
Macfarlane S, Macfarlane GT . (2003). Regulation of short-chain fatty acid production. Proc Nutr Soc 62: 67–72.
Markle JG, Frank DN, Mortin-Toth S, Robertson CE, Feazel LM, Rolle-Kampczyk U et al. (2013). Sex differences in the gut microbiome drive hormone-dependent regulation of autoimmunity. Science 339: 1084–1088.
Mitrou P, Petsiou E, Papakonstantinou E, Maratou E, Lambadiari V, Dimitriadis P et al. (2015). The role of acetic acid on glucose uptake and blood flow rates in the skeletal muscle in humans with impaired glucose tolerance. Eur J Clin Nutr 69: 734–739.
Neu J, Lorca G, Kingma SD, Triplett EW . (2010). The intestinal microbiome: relationship to type 1 diabetes. Endocrinol Metab Clin North Am 39: 563–571.
Pomare EW, Branch WJ, Cummings JH . (1985). Carbohydrate fermentation in the human colon and its relation to acetate concentrations in venous blood. J Clin Invest 75: 1448–1454.
Pozzilli P, Signore A, Williams AJ, Beales PE . (1993). NOD mouse colonies around the world—recent facts and figures. Immunol Today 14: 193–196.
Prochazka M, Serreze DV, Frankel WN, Leiter EH . (1992). NOR/Lt mice: MHC-matched diabetes-resistant control strain for NOD mice. Diabetes 41: 98–106.
Puertollano E, Kolida S, Yaqoob P . (2014). Biological significance of short-chain fatty acid metabolism by the intestinal microbiome. Curr Opin Clin Nutr Metab Care 17: 139–144.
Rabinovitch A, Suarez-Pinzon WL, Sorensen O, Bleackley RC, Power RF . (1995). IFN-gamma gene expression in pancreatic islet-infiltrating mononuclear cells correlates with autoimmune diabetes in nonobese diabetic mice. J Immunol 154: 4874–4882.
Roesch LF, Lorca GL, Casella G, Giongo A, Naranjo A, Pionzio AM et al. (2009). Culture-independent identification of gut bacteria correlated with the onset of diabetes in a rat model. ISME J 3: 536–548.
Sapone A, de Magistris L, Pietzak M, Clemente MG, Tripathi A, Cucca F et al. (2006). Zonulin upregulation is associated with increased gut permeability in subjects with type 1 diabetes and their relatives. Diabetes 55: 1443–1449.
Valladares R, Sankar D, Li N, Williams E, Lai KK, Abdelgeliel AS et al. (2010). Lactobacillus johnsonii N6.2 mitigates the development of type 1 diabetes in BB-DP rats. PLoS One 5: e10507.
Vehik K, Hamman RF, Lezotte D, Norris JM, Klingensmith G, Bloch C et al. (2007). Increasing incidence of type 1 diabetes in 0- to 17-year-old Colorado youth. Diabetes Care 30: 503–509.
Wen L, Ley RE, Volchkov PY, Stranges PB, Avanesyan L, Stonebraker AC et al. (2008). Innate immunity and intestinal microbiota in the development of Type 1 diabetes. Nature 455: 1109–1113.
Yanagibashi T, Hosono A, Oyama A, Tsuda M, Suzuki A, Hachimura S et al. (2013). IgA production in the large intestine is modulated by a different mechanism than in the small intestine: Bacteroides acidifaciens promotes IgA production in the large intestine by inducing germinal center formation and increasing the number of IgA+ B cells. Immunobiology 218: 645–651.
Yurkovetskiy L, Burrows M, Khan AA, Graham L, Volchkov P, Becker L et al. (2013). Gender bias in autoimmunity is influenced by microbiota. Immunity 39: 400–412.
Zhang Y, Lee AS, Shameli A, Geng X, Finegood D, Santamaria P et al. (2010). TLR9 blockade inhibits activation of diabetogenic CD8+ T cells and delays autoimmune diabetes. J Immunol 184: 5645–5653.
Acknowledgements
BAV holds the Children with Intestinal and Liver Disorders (CHILD) Foundation Chair in Pediatric Gastroenterology. BAV is also the Canada Research Chair (Tier 2) in Pediatric Gastroenterology. JPD is a senior scientist at the Child and Family Research Institute. This work was supported by operating grants to BAV from the Canadian Institutes for Health Research, to JPD and BAV from Juvenile Diabetes Research Foundation, to JPD and DLG from the Child and Family Research Institute Diabetes Catalyst Grant from the Canuck’s Foundation and to DLG from VSL#3 Pharmaceuticals and Natural Sciences and Engineering Research Council.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on The ISME Journal website
Rights and permissions
About this article
Cite this article
Brown, K., Godovannyi, A., Ma, C. et al. Prolonged antibiotic treatment induces a diabetogenic intestinal microbiome that accelerates diabetes in NOD mice. ISME J 10, 321–332 (2016). https://doi.org/10.1038/ismej.2015.114
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ismej.2015.114
This article is cited by
-
The potential mechanism of gut microbiota-microbial metabolites-mitochondrial axis in progression of diabetic kidney disease
Molecular Medicine (2023)
-
Complex regulatory effects of gut microbial short-chain fatty acids on immune tolerance and autoimmunity
Cellular & Molecular Immunology (2023)
-
Mechanistic Insights into Immune-Microbiota Interactions and Preventive Role of Probiotics Against Autoimmune Diabetes Mellitus
Probiotics and Antimicrobial Proteins (2023)
-
Microbiome risk profiles as biomarkers for inflammatory and metabolic disorders
Nature Reviews Gastroenterology & Hepatology (2022)
-
Inverse association between use of broad spectrum penicllin with beta-lactamase inhibitors and prevalence of type 1 diabetes mellitus in Europe
Scientific Reports (2021)