Abstract
Disease in oysters has been steadily rising over the past decade, threatening the long-term survival of commercial and natural stocks. Our understanding and management of such diseases are of critical importance as aquaculture is an important aspect of dealing with the approaching worldwide food shortage. Although some bacteria of the Vibrio genus isolated from diseased oysters have been demonstrated to be pathogenic by experimental infection, direct causality has not been established. Little is known about the dynamics of how the bacterial population hosted by oysters changes during disease progression. Combining experimental ecology, a high-throughput infection assay and genome sequencing, we show that the onset of disease in oysters is associated with progressive replacement of diverse benign colonizers by members of a phylogenetically coherent virulent population. Although the virulent population is genetically diverse, all members of that population can cause disease. Comparative genomics across virulent and nonvirulent populations identified candidate virulence factors that were clustered in population-specific genomic regions. Genetic analyses revealed that one gene for a candidate virulent factor, a putative outer membrane protein, is necessary for infection of oysters. Finally, analyses of oyster mortality following experimental infection suggest that disease onset can be facilitated by the presence of nonvirulent strains. This is a new form of polymicrobial disease, in which nonpathogenic strains contribute to increase mortality.
Similar content being viewed by others
Introduction
Vibrios have been associated with successive mortality outbreaks of oyster beds (Crassostrea gigas) in France that have resulted in losses of up to 100% of production (Samain, 2008). Given the near monoculture of C. gigas in Europe, there is an urgent need to understand the epidemiology of these outbreaks, particularly the role of vibrios in the diseases. To date, it has been difficult to determine whether vibrios resident in diseased oysters are mutualistic, opportunistic or pathogenic owing to a lack of diagnostic tools for distinguishing pathogenic from nonpathogenic strains, the fact that individual animals can harbor multiple bacterial genotypes and limitations inherent within the experimental systems available for studying infection. Consequently, it has been difficult to conclusively identify bacterial genotypes or genes that are linked to virulence in oysters.
In the last few years, significant progress has been made in understanding the population structure and diversity of vibrios (Thompson et al., 2005; Hunt et al., 2008). Despite their enormous microdiversity, these organisms fall into well-defined genetic clusters that have similar resource preferences. These clusters have been hypothesized to correspond to populations that act as cohesive ecological units, that is, ecological populations (Hunt et al., 2008). However, a link between ecological populations and pathogenicity has not been demonstrated, and it is unclear whether pathogenicity is a trait primarily linked to clones or to populations comprising a large number of distinct genotypes.
Experimental infections of oysters, which have been performed for a limited number of bacterial strains, have allowed identification of a few factors that contribute to virulence, namely a metalloprotease (Le Roux et al., 2007; Labreuche et al., 2010) and the outer membrane protein OmpU (Duperthuy et al., 2010, 2011). However, knowledge of the absence/presence of these genes is not sufficient for determination of a strain’s pathogenicity (Saulnier et al., 2010). Furthermore, the laboratory analyses previously used to define virulence-linked loci do not capture the complexity of infection within the natural environment. Oysters are typically injected with a single bacterial strain, whereas in their natural environment animals are typically colonized by a diverse assemblage of vibrios (Gay et al., 2004a; Wendling et al., 2014). This diversity may contribute to virulence; in fact, experimental infections have demonstrated that some strains are moderately virulent when injected into animals individually, and display heightened virulence in mixed experimental infections (Gay et al., 2004b).
Oyster vibrioses studied in the laboratory also do not accurately model the natural infection process. Numerous attempts to kill oysters by immersion in vibrio-contaminated sea water have proven unsuccessful, and have necessitated reliance on infection via injection (Gay et al., 2004a; Le Roux et al., 2007; Duperthuy et al., 2011). It is possible that infection in the wild is aided by vibrios’ association with and attachment to other organisms and particles; a recent study has reported that marine aggregates facilitate retention of nanoparticles (including bacteria) by suspension-feeding bivalves (Froelich et al., 2012). Thus, growth of oysters in an environment in which bacteria are not simply in a planktonic form may yield a more accurate understanding of the factors that contribute to virulence.
In the present study, we investigate the oyster disease ecology of microdiverse Vibrio genotypes using a new, field-based approach. We take advantage of recently developed specific pathogen-free spats of C. gigas that become naturally infected when placed in an oceanic environment (Petton et al., 2013). In addition, we use these standardized animals for high-throughput experimental infections. We show that pathogenicity can be ascribed to a cluster of genetically related strains that coincides with a previously defined ecologically cohesive population. Genes specific to this population likely reflect the selective pressure associated with population specialization, and we demonstrate that one of them is required for pathogenicity.
Materials and methods
Strains, plasmid collections and culture conditions
In May 2011, specific pathogen-free oyster spats were transferred to a farming area to allow infection (Supplementary Methods). An oyster set (designed as sentinel) was maintained in the field to monitor the first mortality onset and determine the cumulative mortality rates occurring naturally after 1 month. At the first mortality report, infected animals were reintroduced in the laboratory to reveal the disease. Each day, from a pool of 10 living oysters, vibrios were isolated on selective media (thiosulfate-citrate-bile salts-sucrose agar (TCBS), Difco, BD, Le pont de Claix, France) and re-streaked two times before genotyping using gyrB partial sequence (Supplementary Methods). The strains used for the genomic analyses are described in Table 1. Other bacterial strains are described in Supplementary Table S1. Vibrio isolates were grown in Zobell or Zobell agar, Luria-Bertani (LB) or LB agar (LBA)+NaCl 0.5 M at 20 °C. Escherichia coli strains were grown in LB or on LBA at 37 °C. Chloramphenicol (12 μg ml−1), spectinomycin (100 μg ml−1), thymidine (0.3 mM) and diaminopimelate (0.3 mM) were added as supplements when necessary. Induction of the PBAD promoter was achieved by the addition of 0.2% L-arabinose to the growth media and, conversely, was repressed by the addition of 1% D-glucose.
Genome sequencing, assembly and annotation
A total of 34 strains (Table 1) were sequenced using the Illumina HiSeq 2000 technology (Plateforme de génomique de l'Institut Pasteur, Paris, France) with ∼50-fold coverage (Supplementary Methods). Contigs were assembled de novo using Velvet (Zerbino and Birney, 2008) and genome assembly was improved by contig mapping against the LGP32 reference genome (Le Roux et al., 2009). Computational prediction of coding sequences together with functional assignments were performed using the automated annotation pipeline implemented in the MicroScope platform (Supplementary Methods) (Vallenet et al., 2013).
In silico analyses
A dedicated precomputing repository (marshalling) was created to perform comparative genomic and phylogenomic analyses. Orthologous proteins were defined as reciprocal best hit proteins with 80% MaxLrap and a minimum of 60% identity cutoff (Daubin et al., 2002). The nucleic acid sequences were aligned using Muscle (Edgar, 2004) and filtered by BMGE (Block Mapping and Gathering with Entropy; Criscuolo and Gribaldo, 2010). Phylogenetic trees were built using the parallel version of PhyML applied to maximum-likelihood algorithm and GTR model as parameters (Guindon et al., 2010). A first phylogenetic analysis of concatenated nucleic acid sequences derived from 3229 shared proteins from the 34 genome sequences suggested the clonality of some isolates within our collection (Table 1). This was confirmed by average nucleotide identity value of >99.5% and an accessory genome of <150 coding DNA sequences between isolates. Consequently, only 21/34 isolates were considered as distinct strains.
Vector construction and mutagenesis
Alleles carrying an internal deletion were cloned into a suicide vector using the Gibson method (New England Biolabs, Genopole, Evry, France) (Supplementary Methods). The R6K γ-ori-based suicide vector encodes the ccdB toxin gene under the control of an arabinose-inducible and glucose-repressible promoter, PBAD(Le Roux et al., 2007). Matings between E. coli and Vibrio were performed at 30 °C as described previously (Le Roux et al., 2007) (Supplementary Methods). Selection of the plasmid-borne drug marker (chloramphenicol resistance) resulted in integration of the entire plasmid in the chromosome by a single crossover. Elimination of the plasmid backbone resulting from a second recombination step was selected by arabinose induction of the ccdB toxin gene. Mutants were screened by PCR and are described in Supplementary Table S1. For complementation experiments, the Gibson assembly method was used to clone the R-5.7 gene under a constitutive promoter (PLAC) in pMRB plasmid known to be stable in vibrios (Le Roux et al., 2011). This plasmid was then transferred to Vibrio by conjugation as described previously.
Virulence studies using oysters
Bacteria were grown under constant agitation at 20 °C for 24 h in Zobell media. Then, 100 μl of the culture (106 colony-forming units (CFUs)) pure or diluted were injected intramuscularly into oysters. The bacterial concentration was confirmed by conventional dilution plating on Zobell agar. After injection, the oysters were transferred to aquaria (20 oysters per aquarium of 2.5 l) containing 1 liter of aerated 5 μm filtered sea water at 20 °C, kept under static conditions for 24 h.
Results and discussion
Disease is associated with progressive replacement of nonvirulent vibrios by genetically related virulent strains
Specific pathogen-free oysters were exposed to natural sea water in the field during a mortality outbreak and then returned to the laboratory after 15 days. On each subsequent day, 10 oysters were killed and bacteria were isolated from the tissue (Supplementary Methods). Mortalities started at day 3, reached 50% at day 5 and then ceased (Figure 1a, red bars). The cumulative mortality after 5 days in the laboratory was similar to the extent of mortality observed for a subset of the same batch of oysters maintained in the field for 1 month. We speculate that this mesocosm allows development of disease to proceed more rapidly in the lab.
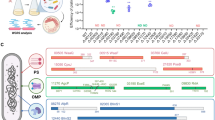
Bacterial population dynamics during oyster infection. (a) Specific pathogen-free oysters were transferred to open sea water for 2 weeks and then transferred to the laboratory to study Vibrio dynamics during the disease expression. Mortalities (pink bar) were recorded daily (D0 to D5) and expressed in percentage (right y axis). D, day. Lines indicate the percentage of strains (left y axis) belonging to the phylogenetic clades described in (b). The black lines correspond to the three most represented clades (clade f: circle; clade d: square; and clade a: triangle) whereas gray lines (clade e: plain; clade c: large dots; and clade b: small dots) correspond to less abundant genotypes. The red line corresponds to the strains inducing >50% mortality (right y axis). (b) Date of isolation and virulence superimposed on the phylogeny of bacterial isolates inferred by maximum-likelihood analysis of partial gyrB gene sequences, with outer and inner rings indicating the % of mortalities obtained 24 h after oyster injection (brown bars >50%; red bars <50%) and the day of isolation (D0 in white, D1–4 in gray gradient and D5 in black, as in (a) below the x axis) respectively. Clades a, b, c, d, e and f were obtained with a bootstrap value of 99%, 94%, 92%, 100%, 100% and 71% and contain the type strains of V. mediterranei, CIP 10320T (clade a), V. chagasii, R-3712T (clade b), V. lentus, CECT 5110T (clade c), V. splendidus, LMG 4042T (clade d), V. cyclitrophicus, LMG 21359T (clade e), V. crassostreae, LGP7T and V. gigantis, LGP13T (clade f). Asterisks indicate the strains sequenced in this study.
Roughly 30 of the bacterial isolates from each day were characterized by partial sequencing of a protein-coding gene (gyrB). Phylogenetic analysis allowed the grouping of 162/173 isolates in 6 clades (designated a to f) with a bootstrap value of >70% (Figure 1b). These clades were matched with named species using type strains V. mediterranei (a), V. chagasii (b), V. lentus (c), V. splendidus (d), V. cyclitrophicus (e), V. crassostreae and V. gigantis (f). Clades b to f belong to the Splendidus super-clade (Sawabe et al., 2013). Strains isolated at the beginning of the experiment were mainly related to clade d (36%) and clade f (39%) (Figure 1a). The clade f strains increased to as high as 77% when the mortalities started but returned to their starting level by the final day. The prevalence of clade d strains declined to 12%, whereas clade a strains increased from 0% to 32% when mortality reached 50%.
To address the pathogenic potential of individual strains, we used an injection model of infection that enables more reliable and rapid infection in the laboratory. Among the 173 isolates individually introduced into specific pathogen-free oysters, 143 (83%) were found to induce <20% mortality (Figure 1b and Supplementary Figure S1). Twenty strains induced >50% mortality and were consequently classified as virulent (vir+). The majority of vir+ strains (75%) clustered into a subclade of (f) that contains the V. crassostreae type strain (LGP7T) (Figure 1b) (Faury et al., 2004). This type strain and other conspecifics were isolated from an oyster mortality event in 2001 and demonstrated to be pathogenic for oysters (Gay et al., 2004a).
The genetic cluster of virulent strains coincides with an ecological population
We investigated whether the vir+/vir− subclades of (f) coincides with any of the ecological populations described by Polz and collaborators (Hunt et al., 2008). As the Vibrio ecological population structure analysis was based on hsp60 sequences, this genetic marker was used to compare the populations (Supplementary Figure S2). The vir+ and vir− strains were found to be included in distinct clades corresponding to ecologically differentiated populations. These two populations were reported to show preferences for either zooplankton or large particles (Hunt et al., 2008).
It is possible that the association between vir+ strains and particles/plankton is important for natural infection of oysters by these pathogens. Recent studies have reported the central role of marine aggregates in facilitating colonization of Crassostrea virginica oysters by Vibrio vulnificus and showed that differences in the ability to incorporate into these aggregates may play a role in the bacterial population disparity observed within oysters (Froelich et al., 2012). Thus, identifying the microhabitats of C. gigas pathogens may facilitate development of an experimental infection model mimicking the natural route of infection, for example, through the use of adapted polymeric substrates. Such approach would also allow for a better understanding of the mode of transmission and primary target tissues or organs for these pathogens.
Comparative genomic analyses reveal limited gene clusters that distinguish vir+ and vir− populations
To identify genetic features that distinguish the vir+ population from vir− strains within clade f, we used high-throughput sequencing (Supplementary Methods) to analyze the genomes of 34 isolates (Figure 1b and Table 1). Of these isolates, 21 appeared to be distinct strains (Table 1): 9 virulent strains (6 from the present study and 3 from a 2001 mortality outbreak) and 12 avirulent strains. The core genome phylogeny (3229 genes) shows that the strains split into two lineages consistent with the pathogenicity status (Figure 2). The average nucleotide identity was 92–93% between populations, whereas within vir+ and vir− lineages, the average nucleotide identity values ranged between 97.8% and –99.9% and between 96.6% and 97.3%, respectively. Despite a strong clonal frame in the core genome, we detected extensive genetic diversity in the flexible genome. Strain-specific gene numbers varied from 1000 to 1700 and from 100 to 1200, respectively, at inter- and intra-population levels, which is two times higher than the flexible genome diversity observed in V. cholerae (Supplementary Figure S3).
Phylogenetic analysis based on concatenated alignments of nucleic acid sequences of core genes of strains sequenced in this study. J2-x and J5-x strains were isolated in this study at days 2 and 5 respectively. LGPx strains were isolated in 2001. LGP7T is the type strain of V. crassostreae species. The strain LGP32 was used as an outgroup. Trees were built by the maximum-likelihood method (GTR substitution model, NNIs, γ4, invariant site) based on sequences aligned using Muscle and filtered with BMGE. Branch lengths are drawn to scale and are proportional to the number of nucleotide changes. Numbers at each node represent the percentage value given by bootstrap analysis of 100 replicates. Strains in which the deletions of the R-5 region were made are underlined.
We performed comparative analyses to identify sequences present in the vir+ and absent from the vir− population. A total of 101 genes were found to be specific to vir+ strains, 53% of which were localized in 7 distinct regions (designated R-1 to 7) (Supplementary Table S2). Four regions (R-2, -3, -4 and -5) are involved in drug resistance and metabolic function, suggesting adaptation of strains to local competitive or environmental pressure. The R-6 region encoding for arylsulfatase may have an important scavenging function in removing sulfate groups from exogenous substrates such as macroalgal polysaccharides and providing carbon sources (Cohen et al., 2007; Mann et al., 2013). The R-1 region is homologous to the widespread colonization island (also named tad gene cluster), which encodes adhesive pili, and was demonstrated as essential for biofilm formation, colonization and pathogenesis in numerous bacteria (Tomich et al., 2007). The R-1 region also encodes the PhoPQ two-component system that controls a variety of processes including resistance to antimicrobial peptides (Otto, 2009). The antimicrobial peptides in concert with reactive oxygen species play a crucial role in the invertebrate immune system (Bachere et al., 2004). Interestingly, in the R-4-specific locus, we identified genes encoding a catalase and a superoxide dismutase putatively implicated in reactive oxygen species resistance (Ibarra and Steele-Mortimer, 2009). Finally, the R-7 region carries genes encoding putative transposases and proteins of unknown function.
Reciprocally, we identified 193 genes present in all vir− strains but absent from all vir+ strains, half of which were localized in 14 regions (Supplementary Table S3). Two regions are putatively involved in phosphonate transport (Yu et al., 2013). Altogether, our data suggest that ecological specialization, possibly through differential association with hosts and/or particulate material, results from gene acquisition conferring function as scavenging, drug resistance, adhesion and host immune response survival.
Clade f vir+ strains encode a putative outer membrane protein that is necessary for virulence
We assessed the importance of vir+-specific loci for V. crassostreae virulence using a genetic knockout approach. Deletion of regions R-1, -2, -4, -5 or -7 in strain J2-9 did not impair bacterial growth in culture media, but deletion of R-5 resulted in a threefold decrease in mortalities induced after bacteria injection (Figure 3a). The importance of R-5 was confirmed using two additional strains (J5-5 and LGP8) belonging to the vir+ population (Figure 3a). Among the 32 genes localized in the R-5 region of J2-9, only 8 genes were present in all vir+ strains and absent from all vir− strains (Figure 3b and Supplementary Table S2). Analyses of mutants lacking individual genes revealed that the R-5.7 gene accounts for the contribution of R-5 to virulence (Figure 3c). When constitutively expressed in trans, R-5.7 was sufficient to restore the virulence of the mutants ΔR5.7 and ΔR5 (Figure 3d). Thus, R-5.7 is the only gene necessary for the contribution of R-5 to virulence. On the other hand, the expression of R-5.7 in trans in a vir− strain was not sufficient to induce a virulent phenotype (Figure 3d). These complementation experiments confirm that R-5.7 gene is necessary but not sufficient for the pathogenicity. The R-5.7 gene (labeled VRSK9J2v1_730268 in J2-9, Supplementary Table S2) is predicted to encode a 798 amino-acid exported protein with a theoretical molecular mass of 89 kDa. No functional domains within the protein could be identified using InterProScan, PFAM or Figfam, but Psort predicted R-5.7 to encode an outer membrane protein. Blast analysis revealed the presence of R-5.7 orthologous genes in several genomes of vibrios belonging to Splendidus, Orientalis and Photobacterium clades (Supplementary Figure S4), although not in LGP32, a V. splendidus-related strain previously demonstrated to be pathogenic for oysters (Le Roux et al., 2009).
Oyster mortality in response to experimental infection with V. crassostreae wild-type (wt) strains and derivatives. A total of 106 CFUs of the tested strains ((a) ΔR-1 to -7 for deleted regions 1 to 7 respectively; (c) ΔR-5.1 to -8 for deleted genes 5.1 to 8 in the region R-5; (d) J2-9 and derivatives ΔR-5, ΔR-5.7 or J2-8 carrying (indicated with a star) or not the expression vector pMRB-PLACR-5.7) were intramuscularly injected into oysters (n=20, in duplicate). Mortality (%) was assessed after 24 h. (b) Organization of the genomic region R-5. Genes in red are found in all vir+ strains and absent in all vir− strains (plain: exported unknown protein; hatched: regulators). Genes in white were also found in vir− strains. Genes in black indicate the region boundaries that have been targeted for the deletion of the entire region (locus tag of the J2-9 strain: VRSK9J2v1_ 73037 and 730270). Arrows indicate genes targeted for single-gene mutagenesis; we were unable to establish a deletion in R-5.4 (gray, italics).
It is notable that of the 81 genes analyzed by deletion only one was found to be necessary for V. crassostreae pathogenicity. This finding suggests that the primary role of these population-specific genes may be unrelated to virulence. It is possible that these shared genes are simply a ‘fossil’ of the common ancestry. On the other hand, the vir+ strains may be specifically adapted to a particular environmental niche where these genes are beneficial. In such a scheme, oysters may be considered as an alternative habitat for the vir+ population rather than their principal environmental niche. This hypothesis is in accordance with previous data demonstrating that V. crassostreae was associated with both algal detritus and zooplankton (Preheim et al., 2011; Szabo et al., 2012). Further mapping of virulent strains onto V. crassostreae ecological populations may enable determination of the microhabitats from which oyster pathogens emerge and provide more insight into the nature of populations that serve as reservoirs of pathogens.
Nonvirulent strains may facilitate the disease
Naturally infected oysters initially contain a large proportion of avirulent strains, but these are progressively replaced by a virulent population that comprises ∼50% of the bacterial isolates at the point of maximal mortalities (Figure 1a). The low prevalence of vir+ strains in the early infection process could reflect a contribution of the nonvirulent strains to the development of disease. To address this question, the vir+ strain J2-9 (V. crassostreae) was injected into oysters at various doses, either alone or in combination with the nonvirulent strains J2-8 (clade f) or J2-20 (Shewanella sp.) (Figure 4). When injected alone, reduction of the injected dose of J2-9 from 106 to 4 × 104 CFUs (via dilution in culture media) significantly reduced oyster mortality (∼90% vs 5% mortality), even if the infection was allowed to progress for a longer time. In contrast, when J2-9 was injected at 4 × 104 CFUs following serial dilutions with pure cultures of vir- strains (J2-8 or J2-20), so that the final CFUs per inoculum were 106, mortality rates were markedly higher than with J2-9 alone (cumulative mortalities of 70% for J2-9 diluted in J2-8 and 60% for J2-9 diluted in J2-20). Thus, the presence of nonvirulent bacteria dramatically increases the virulence of low doses of J2-9, suggesting that there are genotype-independent effects of bacterial density upon virulence.
Oyster mortality in response to experimental infection with vir+ strain in the presence of vir− strain cultures. The J2-9 (V. crassostreae, vir+) was intramuscularly injected into oysters (two aquaria of n=20) in pure culture (106 CFUs per animal), diluted (4 × 104 CFUs per animal) with culture media or with a pure culture of vir− strains J2-8 (clade f, vir−) or J2-20 (Shewanella sp.). As a negative control, a pure culture (106 CFUs per animal) of J2-8 or J2-20 was also injected. Mortality (%) was assessed after 24 h.
Thus, although vir− strains are not sufficient for pathogenesis, they clearly have some features (as yet undetermined) that contribute either directly or indirectly to virulence. One possibility is that vir− strains provide resources lacking by the vir+, enabling the vir+ strains to act as ‘cheaters’, as seen in some analyses of siderophore synthesis and utilization (Cordero et al., 2012). An alternate role for the vir− strains may be to generate a sufficient bacterial load, either to overcome host defenses or to induce expression of virulence factors that are regulated by quorum sensing (Bassler, 2002). Notably, autoinducer synthases (CsqA, LuxM and LuxS), which initiate the quorum sensing signaling cascades, appear to be encoded by both vir+ and vir− strains. In the future, we will investigate the importance of quorum sensing pathways in virulence, as well as explore additional means by which vir− strains contribute to the disease in order to better understand this process of density-dependent pathogenesis.
Conclusion
Our results demonstrate the consistency of the virulent population that also corresponds to a previously identified ecologically cohesive genotypic cluster. In the future, delineation of ecological populations together with experimental infections should allow the determination of populations with high or low risk of pathogenicity, the microhabitats from which oyster pathogens emerge and, consequently, which populations serve as reservoirs of pathogens. Hence, it may be possible to develop diagnostic tools at the taxonomic level as soon as population-specific genes are targeted.
References
Bachere E, Gueguen Y, Gonzalez M, de Lorgeril J, Garnier J, Romestand B . (2004). Insights into the anti-microbial defense of marine invertebrates: the penaeid shrimps and the oyster Crassostrea gigas. Immunol Rev 198: 149–168.
Bassler BL . (2002). Small talk. Cell-to-cell communication in bacteria. Cell 109: 421–424.
Cohen AL, Oliver JD, DePaola A, Feil EJ, Boyd EF . (2007). Emergence of a virulent clade of Vibrio vulnificus and correlation with the presence of a 33-kilobase genomic island. Appl Environ Microbiol 73: 5553–5565.
Cordero OX, Wildschutte H, Kirkup B, Proehl S, Ngo L, Hussain F et al. (2012). Ecological populations of bacteria act as socially cohesive units of antibiotic production and resistance. Science 337: 1228–1231.
Criscuolo A, Gribaldo S . (2010). BMGE (Block Mapping and Gathering with Entropy): a new software for selection of phylogenetic informative regions from multiple sequence alignments. BMC Evol Biol 10: 210.
Daubin V, Gouy M, Perriere G . (2002). A phylogenomic approach to bacterial phylogeny: evidence of a core of genes sharing a common history. Genome Res 12: 1080–1090.
Duperthuy M, Binesse J, Le Roux F, Romestand B, Caro A, Got P et al. (2010). The major outer membrane protein OmpU of Vibrio splendidus contributes to host antimicrobial peptide resistance and is required for virulence in the oyster Crassostrea gigas. Environ Microbiol 12: 951–963.
Duperthuy M, Schmitt P, Garzon E, Caro A, Rosa RD, Le Roux F et al. (2011). Use of OmpU porins for attachment and invasion of Crassostrea gigas immune cells by the oyster pathogen Vibrio splendidus. Proc Natl Acad Sci USA 108: 2993–2998.
Edgar RC . (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32: 1792–1797.
Faury N, Saulnier D, Thompson FL, Gay M, Swings J, Le Roux F . (2004). Vibrio crassostreae sp. nov., isolated from the haemolymph of oysters (Crassostrea gigas). Int J Syst Evol Microbiol 54: 2137–2140.
Froelich B, Ayrapetyan M, Oliver JD . (2012). Vibrio vulnificus integration into marine aggregates and subsequent uptake by the oyster, Crassostrea virginica. Appl Environ Microbiol 79: 1454–1458.
Gay M, Berthe FC, Le Roux F . (2004a). Screening of Vibrio isolates to develop an experimental infection model in the Pacific oyster Crassostrea gigas. Dis Aquat Organ 59: 49–56.
Gay M, Renault T, Pons AM, Le Roux F . (2004b). Two Vibrio splendidus related strains collaborate to kill Crassostrea gigas: taxonomy and host alterations. Dis Aquat Organ 62: 65–74.
Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O . (2010). New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol 59: 307–321.
Hunt DE, David LA, Gevers D, Preheim SP, Alm EJ, Polz MF . (2008). Resource partitioning and sympatric differentiation among closely related bacterioplankton. Science 320: 1081–1085.
Ibarra JA, Steele-Mortimer O . (2009). Salmonella—the ultimate insider. Salmonella virulence factors that modulate intracellular survival. Cell Microbiol 11: 1579–1586.
Labreuche Y, Le Roux F, Henry J, Zatylny C, Huvet A, Lambert C et al. (2010). Vibrio aestuarianus zinc metalloprotease causes lethality in the Pacific oyster Crassostrea gigas and impairs the host cellular immune defenses. Fish Shellfish Immunol 29: 753–758.
Le Roux F, Binesse J, Saulnier D, Mazel D . (2007). Construction of a Vibrio splendidus mutant lacking the metalloprotease gene vsm by use of a novel counterselectable suicide vector. Appl Environ Microbiol 73: 777–784.
Le Roux F, Zouine M, Chakroun N, Binesse J, Saulnier D, Bouchier C et al. (2009). Genome sequence of Vibrio splendidus: an abundant planctonic marine species with a large genotypic diversity. Environ Microbiol 11: 1959–1970.
Le Roux F, Davis BM, Waldor MK . (2011). Conserved small RNAs govern replication and incompatibility of a diverse new plasmid family from marine bacteria. Nucleic Acids Res 39: 1004–1013.
Mann AJ, Hahnke RL, Huang S, Werner J, Xing P, Barbeyron T et al. (2013). The genome of the alga-associated marine flavobacterium Formosa agariphila KMM 3901T reveals a broad potential for degradation of algal polysaccharides. Appl Environ Microbiol 79: 6813–6822.
Otto M . (2009). Bacterial sensing of antimicrobial peptides. Contrib Microbiol 16: 136–149.
Petton B, Pernet F, Robert R, Boudry P . (2013). Temperature influence on pathogen transmission and subsequent mortalities in juvenile Pacific oysters Crassostrea gigas. Aquacult Environ Interact 3: 257–273.
Preheim SP, Timberlake S, Polz MF . (2011). Merging taxonomy with ecological population prediction in a case study of Vibrionaceae. Appl Environ Microbiol 77: 7195–7206.
Samain JF . (2008). Summer mortality of Pacific oyster Crassostrea gigas. The Morest Project, Versailles, Editions Quae, 37p.
Saulnier D, De Decker S, Haffner P, Cobret L, Robert M, Garcia C . (2010). A large-scale epidemiological study to identify bacteria pathogenic to Pacific oyster Crassostrea gigas and correlation between virulence and metalloprotease-like activity. Microbial Ecol 59: 787–798.
Sawabe T, Ogura Y, Matsumura Y, Feng G, Amin AR, Mino S et al. (2013). Updating the Vibrio clades defined by multilocus sequence phylogeny: proposal of eight new clades, and the description of Vibrio tritonius sp. nov. Front Microbiol 4: 414.
Szabo G, Preheim SP, Kauffman KM, David LA, Shapiro J, Alm EJ et al. (2012). Reproducibility of Vibrionaceae population structure in coastal bacterioplankton. ISME J 7: 509–519.
Thompson JR, Pacocha S, Pharino C, Klepac-Ceraj V, Hunt DE, Benoit J et al. (2005). Genotypic diversity within a natural coastal bacterioplankton population. Science 307: 1311–1313.
Tomich M, Planet PJ, Figurski DH . (2007). The tad locus: postcards from the widespread colonization island. Nat Rev Microbiol 5: 363–375.
Vallenet D, Belda E, Calteau A, Cruveiller S, Engelen S, Lajus A et al. (2013). MicroScope—an integrated microbial resource for the curation and comparative analysis of genomic and metabolic data. Nucleic Acids Res 41: D636–D647.
Wendling CC, Batista FM, Wegner KM . (2014). Persistence, seasonal dynamics and pathogenic potential of Vibrio communities from pacific oyster hemolymph. PLoS One 9: e94256.
Yu X, Doroghazi JR, Janga SC, Zhang JK, Circello B, Griffin BM et al. (2013). Diversity and abundance of phosphonate biosynthetic genes in nature. Proc Natl Acad Sci USA 110: 20759–20764.
Zerbino DR, Birney E . (2008). Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res 18: 821–829.
Acknowledgements
We warmly thank Brigid Davis (HMS, Boston, USA), Marylise Duperthuy (Umea University, Sweden), Otto X Cordero (ETH Zurich, CH) and Martin F Polz (MIT, Cambridge, USA) for fruitful discussions and critically reading the manuscript. We thank the staff of the station Ifremer Argenton and Bouin, the ABIMS and CRBM (Roscoff) and LABGeM (Evry) platforms for technical support. FLR acknowledges the VIBRIOGEN consortium (Annick Jacq, Delphine Destoumieux and Guillaume Charrière) for our precious exchanges. This study has been supported by the Region Bretagne (SAD Vibrigen 6633; to TV), the ANR blanc (11-BSV7-023-01 ‘VIBRIOGEN’; to DG), the EMBRC France (to AL) and Ifremer (to DG and TV).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Author Contributions
AL, TV, BP, YL and FLR performed experiments; DG and AC performed the in silico analyses; YL and FLR designed experiments, interpreted results and wrote the paper; AL, DG and TV contribute equally to this work. The manuscript has been seen and approved by all authors. The material represents an original result and has not been submitted for publication elsewhere.
Supplementary Information accompanies this paper on The ISME Journal website
Supplementary information
Rights and permissions
About this article
Cite this article
Lemire, A., Goudenège, D., Versigny, T. et al. Populations, not clones, are the unit of vibrio pathogenesis in naturally infected oysters. ISME J 9, 1523–1531 (2015). https://doi.org/10.1038/ismej.2014.233
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ismej.2014.233
This article is cited by
-
A core of functional complementary bacteria infects oysters in Pacific Oyster Mortality Syndrome
Animal Microbiome (2023)
-
A MALDI-TOF MS database for fast identification of Vibrio spp. potentially pathogenic to marine mollusks
Applied Microbiology and Biotechnology (2021)
-
Oyster hemolymph is a complex and dynamic ecosystem hosting bacteria, protists and viruses
Animal Microbiome (2020)
-
Achieving a multi-strain symbiosis: strain behavior and infection dynamics
The ISME Journal (2019)
-
Characterisation of the Pacific Oyster Microbiome During a Summer Mortality Event
Microbial Ecology (2019)