Abstract
Ascidians contain abundant, diverse secondary metabolites, which are thought to serve a defensive role and which have been applied to drug discovery. It is known that bacteria in symbiosis with ascidians produce several of these metabolites, but very little is known about factors governing these ‘chemical symbioses’. To examine this phenomenon across a wide geographical and species scale, we performed bacterial and chemical analyses of 32 different ascidians, mostly from the didemnid family from Florida, Southern California and a broad expanse of the tropical Pacific Ocean. Bacterial diversity analysis showed that ascidian microbiomes are highly diverse, and this diversity does not correlate with geographical location or latitude. Within a subset of species, ascidian microbiomes are also stable over time (R=−0.037, P-value=0.499). Ascidian microbiomes and metabolomes contain species-specific and location-specific components. Location-specific bacteria are found in low abundance in the ascidians and mostly represent strains that are widespread. Location-specific metabolites consist largely of lipids, which may reflect differences in water temperature. By contrast, species-specific bacteria are mostly abundant sequenced components of the microbiomes and include secondary metabolite producers as major components. Species-specific chemicals are dominated by secondary metabolites. Together with previous analyses that focused on single ascidian species or symbiont type, these results reveal fundamental properties of secondary metabolic symbiosis. Different ascidian species have established associations with many different bacterial symbionts, including those known to produce toxic chemicals. This implies a strong selection for this property and the independent origin of secondary metabolite-based associations in different ascidian species. The analysis here streamlines the connection of secondary metabolite to producing bacterium, enabling further biological and biotechnological studies.
Similar content being viewed by others
Introduction
Filter-feeding chordates known as ascidians are found in all of the world’s oceans (Shenkar and Swalla, 2011; Schmidt et al., 2012). Ascidians are sessile, soft-bodied animals that are vulnerable to predation. Many ascidian species contain abundant, potently toxic secondary metabolites that are implicated in chemical defense. Indeed, there are several examples in which a defensive role has been experimentally assigned to individual secondary metabolites (Paul et al., 1990; Joullie et al., 2003). Strikingly, there are now several examples in which it has been shown that symbiotic bacteria produce potentially defensive metabolites, while the host animals do not (Supplementary Table S8) (Schmidt et al., 2005; Donia et al., 2006, 2008; Rath et al., 2011; Donia et al., 2011b, 2011c; Kwan et al., 2012). Beyond defense, symbiotic metabolites in ascidians are also known to have other roles. For example, mycosporine amino acids are critical to the defense of tropical ascidians against ultraviolet irradiation, but they are produced by symbiotic bacteria (Hirose et al., 2006; Donia et al., 2011a). Thus, symbiotic bacteria contribute critical secondary metabolites that are apparently necessary for the survival of the animals.
In tropical ascidians of the Family Didemnidae, Prochloron spp. symbiotic bacteria produce a variety of toxic, cyclic peptides known as cyanobactins (Donia et al., 2008), as well as ultraviolet-protectant mycosporine amino acids (Donia et al., 2011a). Prochloron seems to be obligate for the host animals, yet horizontal transmission dominates for this group (Hirose and Fukuda, 2006; Hirose et al., 2009; Kojima and Hirose, 2010). Nonetheless, isolated Prochloron chemistry is strongly ascidian species specific, implicating a chemical driver in the interaction (Donia et al., 2011b). By contrast, certain groups of highly toxic polyketides are synthesized by genome-reduced, obligate intracellular alpha-proteobacteria, Candidatus Endolissoclinum faulkneri (Kwan et al., 2012). This interaction has resulted in vertical transmission and co-speciation that has been ongoing for approximately 6–35 million years (Kwan et al., 2012; Kwan and Schmidt, 2013). During that period, the production of toxic secondary metabolism has been maintained while the remainder of the symbiont’s genome has degraded. Another proteobacterium, the gamma-proteobacterium Candidatus Endoecteinascidia frumentensis, produces potent ecteinascidins within the non-didemnid ascidian, Ecteinascidia turbinata (Rath et al., 2011). Similarly, correlative evidence suggests that alpha-proteobacteria may produce polyketides in an Antarctic didemnid ascidian (Riesenfeld et al., 2008). Finally, indirect but compelling evidence implicates symbiotic bacteria in the production of a variety of other ascidian metabolites (Schmidt and Donia, 2010; Xu et al., 2012).
Although symbiotic bacteria have been shown to produce about 7% of known ascidian metabolites, the source of about 93% of described metabolites remains unknown. Additionally, it is probable that most ascidian metabolites remain undescribed. Here, we sought to examine the factors underlying production of secondary metabolites in ascidians globally, with the idea that this would be useful in rapidly identifying new bioactive natural products and in determining the true producers of ascidian compounds. We compared the microbiomes and metabolomes of ascidians from tropical, subtropical and temperate waters. We broadly studied the link between symbiotic bacteria and the presence of secondary metabolites. The long-term outcome of these studies will help establish an understanding of how symbiotic bacterial communities change and evolve across ascidian phylogeny as well as in time and space. Developing this big picture view provides a basis for collection strategies as well as increase our understanding of what factors might affect symbioses and natural products in ascidians.
Materials and methods
Sample collection and processing
Ascidian samples were collected by scuba diving and processed in the field, except for samples from Florida, which were shipped live by a professional collector. Each ascidian sample was divided into fractions that were preserved in RNAlater (Ambion, Austin, TX, USA) for the microbiome analyses and fractions that were frozen for the metabolomic analyses. In this study, we used samples collected from different locations (year of collection): Papua New Guinea (2011); Fiji (2006); Vanuatu (2008); Catalina Island, California (2010 and 2011); and Florida Keys (2011) (see Supplementary Table S1 for sampling details).
18S rRNA gene sequence analysis
Ascidian 18S rRNA genes were amplified from the same genomic DNA used for 16S rRNA gene sequencing. In cases where amplification was challenging, genomic DNA was extracted using an alternative method (Sokolov, 2000). 18S rRNA genes were amplified using combinations of the following: forward primers AscF3 and AscF1, reverse primers AscR1 and AscR3 (Yokobori et al., 2006) and AscF2 and AscR5 (this study) (Supplementary Table S3). The amplicons were submitted for direct sequencing, leading to successful analysis of 17 out of the 32 samples. For the remaining 15, PCR products were cloned into Topo TA Zero blunt vector (Invitrogen, Carlsbad, CA, USA), and individual clones were sequenced. Primers AscF2 and AscR5 were used for samples that showed non-ascidian 18S products after the first round of sequencing (primers used for each sample are defined in Supplementary Table S1). The ascidian sequences obtained were aligned using MUSCLE (Edgar, 2004) and trimmed using Gblocks (Castresana, 2000). The parameters used for Gblocks were the following: minimum number of sequences for a conserved position=25, minimum number of sequences for a flanking position=40, minimum number for a contiguous non-conserved position=8, minimum length of a block=10, allowed gap position=none. Of the 2013 original positions, 739 (36%) were retained and used for construction of the phylogenetic tree. The 18S tree was constructed using FastTree MP (Price et al., 2010) and visualized using the Interactive Tree of Life server (Letunic and Bork, 2011). Additional ascidian sequences included in the tree were retrieved from GenBank (see Supplementary Figure S1 for accession numbers). 18S sequences from this data set were deposited in Genbank under accession numbers KJ720704–KJ720729, KJ009376 (L. patella Fiji), KJ009378 (L. patella PNG_11040), KJ009377 (L. patella PNG_11033).
DNA extraction and preparation
Metagenomic DNA was extracted using previous methods (Schmidt and Donia, 2009) with modifications (see Supplementary Methods for details). The DNA obtained was further purified through either Genomic Tip (Qiagen, Valencia, CA, USA) or Genomic DNA Clean and Concentrator (Zymo Research, Irvine, CA, USA) kits and quantified using a NanoDrop spectrophotometer (Thermo Scientific, Waltham, MA, USA). The quality of DNA was checked by amplifying 16S rRNA gene PCR using 24F and 1492R primers (Supplementary Table S3). Samples that passed the test were used for 16S rRNA gene amplification and pyrosequencing at Research and Testing Laboratory (RTL, Lubbock, TX, USA). RTL amplified a portion of the 16S rRNA gene using primer forward primer 939F and reverse primer 1492R (Supplementary Table S3) spanning the V6–V9 hypervariable regions in the bacterial 16S rRNA gene and sequenced the resulting amplicons by 454 (Roche, Branford, CT, USA).
16S rRNA gene sequence analysis
After sequencing, the reads were denoised and checked for chimeras (UCHIME; Edgar et al., 2011). Reads were then quality filtered, and the RTL-generated FASTA files with the corresponding quality scores and mapping files were demultiplexed in the QIIME analysis pipeline (Caporaso et al., 2010b). All ascidian samples analyzed in this study were then joined together for operational taxonomic unit (OTU) picking, taxonomy assignment and further diversity analyses in QIIME. OTUs were assigned at 97% identity, corresponding approximately to a species-level assignment for bacteria. A representative set of UCLUST (Edgar, 2010) picked OTUs were aligned using PyNAST (Caporaso et al., 2010a) and passed through the RDP classifier using the Greengenes reference sequences (version 2_10) (DeSantis et al., 2006; Wang et al., 2007). The BIOME or table format output from this analysis was then used as input for downstream analyses in QIIME and R (McMurdie and Holmes, 2013; Oksanen et al., 2013; R Core Team, 2013). The analysis was done in parallel on Clovr 16S microbial diversity pipeline for comparison (Angiuoli et al., 2011). Alpha and beta diversity analyses were performed within QIIME using alpha_rarefaction.py (parameters included the Phylogenetic Diversity (PD), Shannon, Simpson and Observed Species indices) and beta_diversity_through_plots.py, respectively. Principal coordinates analysis was then performed using the UniFrac distance matrix (Lozupone et al., 2011) and visualized in R. Hierarchical clustering and heatmaps were done in R. For networking analysis, categories including location and different host categories (see below) were included in the mapping file. G tests for independence were done to test whether samples are more connected within these categories than expected by chance. Visualization of the networks was performed in Cytoscape (Kohl et al., 2011) using the edge-weighted spring-embedded layout. Analysis of similarity (ANOSIM) calculations were performed to compare microbiome diversity across species and within species across time and location (Clarke, 1993; Chapman and Underwood, 1999). ANOSIM is a distribution-free analysis of variance method that relies on calculating a distance metric (we used Bray–Curtis dissimilarity values) and then comparing the difference between within- and between-sample distances using a permutation test approach (we required a minimum of two samples/group and used 100 000 permutations) (Clarke, 1993). A sensitivity analysis was performed using a similar method (‘adonis’ function) also available in the vegan R package. To compare the diversity measures between location and hosts, we used a permutation sampling test method that was developed specifically for comparing diversity measures (Pallmann et al., 2012). The tests were performed comparing each host or location to the overall mean diversity (GrandMean option) using the mcpHill function in the SimBoot R package for locations and hosts with a minimum of two samples (Pallmann et al., 2012). P-values were adjusted for multiple comparisons across groups (comparing each group with the overall mean) and across diversity measures (Shannon and Simpson). These sequence data have been submitted to SRA, under BioProjects accession SAMN02693397–SAMN02693428.
Ultra High Pressure Liquid Chromatography (LC)/high-resolution mass spectrometry (MS) analysis
LC/MS data (see Supplementary Methods for sample preparation) were acquired using a Bruker MaXis electrospray ionization quadrupole time-of-flight (ESI-Q-TOF) mass spectrometer (Bruker, Billerica, MA, USA) coupled with a Waters Acquity UPLC system (Waters, Millford, MA, USA) operated by the Bruker Hystar software (Bruker). A gradient of methanol and H2O (containing 0.1% formic acid) was used with a flow rate of 0.3 ml min−1 on a RP C18 column (Phenomenex Kinetex 2.6 μm, 2.1 × 100 mm, Phenomenex, Torrance, CA, USA). The gradient went from 10% methanol/90% H2O to 97% methanol/3% H2O in 12 min, followed by 97% methanol/3% H2O held for 3.5 min. Full-scan mass spectra (m/z 150–1550) were measured in the positive ESI mode. The mass spectrometer was operated using previously published parameters (Hou et al., 2012). Tune mix (Agilent Technologies, Santa Clara, CA, USA; ESI-L low concentration) was introduced through a divert valve at the end of each chromatographic run for automatic internal calibration. Data-dependent MS/MS were acquired as previously published (Hou et al., 2012).
LC/MS data processing and compound identification
LC/MS data was bucketed and Principal Component Analysis (PCA) was performed using Bruker ProfileAnalysis 2.0. The following parameters were applied to find molecular features: S/N threshold, 5; correlation coefficient threshold, 0.7; minimum compound length, 10 spectra; and smoothing width, 1. Bucket generation was performed in the retention time range 60–930 s and in a mass range from m/z 200 to 1500. All other bucket generation parameters were the same as previously published (Hou et al., 2012). For further PCA analysis, the buckets obtained were filtered to intensities >1.0e4 as the threshold determined after inspection of individual chromatograms. PCA was then performed in R using the prcomp() function. The PCA biplot generated was used as a preliminary guide for identification of extract components that are correlated with certain clustering patterns of ascidian samples. For graphing and further clustering, principal components were chosen based on their eigenvalues or based on their contribution to the cumulative variance (>80%). Hierarchical clustering was performed in R using the Euclidean distance between the extracted values. For compound identification, mzxml files from LC/MS were processed in the MzMine software (Pluskal et al., 2010).
Results
Phylogeny and diversity of ascidian samples
We examined the microbiomes and chemistry of 32 ascidian samples collected from four locations representing the temperate (Catalina Island, California), subtropical (Florida Keys), and tropical (Papua New Guinea, Fiji and Vanuatu) ocean regions at different times during the period 2006–2011 (Figure 1). We chose samples from the Family Didemnidae plus a few other representative sets from other ascidian families. Phylogenetic analysis using the 18S rRNA gene sequences confirmed that most of the samples are didemnid ascidians (21 samples) from the order Aplousobranchia (26 samples). Representative samples from the order Phlebobranchia (three individual samples) and Stolidobranchia (three samples) were also obtained (Supplementary Figure S1). Samples from the order Aplousobranchia include Eudistoma sp. (2 individuals), Cystodytes sp. (3 individuals), Lissoclinum sp. (9 individuals), Trididemnum sp. (2 individuals) and Didemnum sp. (10 individuals). Stolidobranchia samples include Botryllus (1), Styela (1) and Pyura (1) spp., while Ecteinascidia (2) and Ascidia (1) spp. represent the order Phlebobranchia.
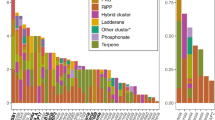
Collection sites and conception of this study. Samples were obtained from a wide variety of geographical locations (triangles). A subset of samples contained natural products where the producing symbiotic bacteria were already known (boxes). Also shown in boxes are phylogenetic trees of the top-10 bacteria found in each sample (see Supplementary Figure S2C for details). Within these top 10, the natural product-producing symbiont is a major representative. This information was compared with metabolomic and microbiome analysis from ascidians where the natural products and producing organisms are not known, with the purpose of understanding the global relationship between natural products and symbiotic bacteria in ascidians.
In the course of sequencing 18S rRNA genes, we were also able to identify other eukaryotes that were associated with the ascidian tissues. These were obtained from many trials using different sets of 18S primers (Supplementary Table S3, see Methods for details). For example, we identified the crustacean species Notodelphys prasina, which were consistently associated with the Cystodytes sp. samples. Indeed, these crustaceans have been reported as seasonal parasites of Cystodytes sp. (Monniot et al., 1991). Nematodes, apicomplexans, copepods and other uncultured eukaryotes are also among the organisms identified from the 18S rRNA gene sequencing (Supplementary Table S7).
Bacterial diversity
Using barcoded 16S rRNA gene sequencing (Roche 454), we obtained an average of 6784 sequences per sample, with a total of 217 703 reads from all ascidian samples. With an OTU defined as >97% 16S rRNA gene sequence identity, we observed a total of 3892 OTUs (10–804 OTUs per sample), indicating a rich and diverse bacterial population (Supplementary Table S2). Using the Shannon index and the other diversity measures, alpha diversity was compared according to variables such as location, ascidian host species and season (Figures 2a and b; Supplementary Table S4). We had hypothesized that there may be a difference in diversity by latitude and that this might in part explain the more abundant secondary metabolism ascribed to tropical ascidians. However, from the rarefaction curves there was no apparent correlation between the diversity of the microbiomes and geographical location (Figure 2a). We further confirmed this by showing that there was no statistical difference in the diversity between locations or latitudinal regions (CA, FL, PNG/FJ/VN; mcpHill analysis using the Shannon index: P=0.950–0.990, Supplementary Figure S9). We also compared the diversity between host species in groups where we had at least two samples per species. There was also no statistical difference in diversity between the other species (P=0.600–1.000, Supplementary Figure S9), except for a group of Didemnum sp. from Papua New Guinea (Didemnum sp.2, Supplementary Figure S9), which had a higher diversity compared with the rest of the groups (P=0.008). Results were similar for the Simpson diversity analysis. Two individual samples from this data set exhibited distinctly low diversity: Eudistoma sp. (41 OTUs of 13886 reads) and Trididemnum sp. (10 OTUs of 572 reads). More samples would be needed to determine whether these species are actually less diverse.
Host specificity of ascidian microbiomes
We aimed to determine which factors (host phylogeny, species identity, location, time/water temperature) were related to microbiome composition in ascidians. We first used two types of cluster analyses: networks (based upon OTU) and UniFrac (based upon phylogeny). A significant clustering by host species was observed from the network-based analysis (Figure 3a) (G=4.990, P=0.010). This was clearly supported by the dendrogram subclusters from the weighted Unifrac analysis in cases where we had multiple samples of the same hosts (Figure 3b). We further analyzed the differences in the community structure of the microbiomes between host species and found a statistically significant difference in the microbiomes across species collected in the same location at the same time (11 samples from 3 species were compared, ANOSIM: R=0.762, P=0.002, Supplementary Table S9). These results indicated that a major component of the ascidian microbiome was host specific and did not merely result from transient interactions with seawater or other environmental factors.
Host specificity of ascidian microbiomes assessed by UniFrac and network-based analyses. (a) Cytoscape network of ascidian microbiomes showing a significant clustering by host species. Categories are reflected in the color scheme, where identical species (defined in map 1 of Supplementary Table S5) have identical colors. (b) Unifrac-based hierarchical clustering of ascidians plotted in two symmetrical dimensions. Groups at the right most portion indicate the grouping used for the analysis of species-specific components of the microbiomes.
We further examined host specificity by testing microbiome stability over time and space. We compared a subset of ascidians from California that were collected in the spring (CA_Didemnum_sp_4.2 and CA_Didemnum_sp_4.17, CA_Didemnum_sp_4.7) and fall (CA_Didemnum_sp_5.1, CA_Didemnum_sp_5.11, and CA_Didemnum_sp_5.18) of 2011. There were no significant seasonal differences in the two sample sets (ANOSIM: R=−0.037, P=0.490), suggesting the existence of a stable and host-specific microbiome over time. The lack of a change in the microbiome with time and space is further supported by the analysis of L. patella samples from the tropical Pacific. These samples were collected at widely different times (different years) and places, and yet they consistently clustered with a similar microbiome across time and space and grouped differently than other species collected at the same locations (Figure 3b). This finding is also consistent with recent reports of ascidian microbiomes, in which some of the same ascidian species we report here were previously examined in other locations (Erwin et al., 2013; Dishaw et al., 2014). Consistently similar bacteria are found across time and space in these studies, in an ascidian host-specific manner.
If the microbiome is intrinsic to the seawater rather than to ascidians, one would expect that ascidians sampled at a particular time and place would have similar microbiomes. Across samples from different ascidian species collected in California or Papua New Guinea, there was a statistically significant difference in microbiome types (CA samples P=0.005, PNG samples P=0.04). Across samples within a single ascidian species collected at these locations, there was no statistically significant difference in microbiome types (P>0.05), as expected (Supplementary Table S9). Taken together with the analysis described above, these results indicate that there is an ascidian species-specific microbiome, which appears to be stable across time and space, within the limits of samples studied so far. Below, we show that these species-specific bacterial sequences represent the most abundant (usually top 10) sequences present in the samples.
Geographical component of ascidian microbiomes
In addition to the host-specific signal, we also observed a significant grouping of the ascidian microbiomes by geographical location or latitudinal region (Figure 4a; G=6.22, P=0.006). This at first seemed contradictory to the findings of host specificity, prompting us to perform further analysis. In so doing, we found that the location-specific component of the microbiome differed from that found in the host-specific component. As further described below, the location-specific component includes the low-abundance sequences found in the microbiomes. It likely reflects the environment of the ascidians, including the seawater in which they live.
Cytoscape networks of ascidians evaluated using different categories of hosts and location. (a) Significant clustering of ascidians according to location, where each color indicates a location. (b) Ascidians are still clustered by location even after redefining the host species grouping at different levels and excluding ascidian groups that had only been collected at the same location. The grouping shown here is defined in map 3 of Supplementary Table S5.
We first hypothesized that the location-specific component was due to the overlap between the ascidian species and location in our sample set, as some species were all collected from the same location (for example, Cystodytes sp., L. badium, E. turbinata). As an alternative hypothesis, because we used the 18S rRNA gene as the marker for phylogeny, we considered that the host speciation might not be optimally resolved. To address these possibilities, we repeated the analyses by defining host species at different levels (Supplementary Table S5) according to their position in the 18S phylogenetic tree and by excluding the ascidian groups that were only collected from a single location (Supplementary Table S5, Figure 4b). Despite some statistical differences (changes in G tests), the geographical signals for the microbiomes were maintained, disproving the hypotheses and indicating a likely geographical component of ascidian microbiomes. This was further examined by determining which microbial groups were generally shared by most samples from those locations regardless of species. The geographical component was comprised of relatively rare sequences in the ascidian microbiome (described below). These sequences mostly represented relatively commonly sequenced bacterial strains found in seawater, and thus they may, at least in part, represent a component of the microbiome derived from local seawater. Many of these strains are the same as those found in other ascidian microbiomes, so that even if they are derived from the environment they are still largely associated with ascidians. This might be due to physical factors (filtration particle size; digestion efficiency in ascidians; and so on) or to biological factors, such as an actual symbiotic association between the organisms.
Bacterial composition of ascidians
We identified 18 known bacterial phyla, 16 candidate bacterial phyla and an unclassified bacterial group comprising 285 OTUs (Figure 2c). The most abundant phylum is Proteobacteria (2074 OTUs), comprising more than half of the total number of OTUs from all ascidian samples. Proteobacteria is the most abundant phylum in all samples with the exceptions of L. patella, L. bistratum and Styela plicata, in which Cyanobacteria is the most abundant group. We also detected other phyla, including Acidobacteria, Nitrospira and Chloroflexi, in relatively minor amounts in the samples, as well as other rare and minor (<1% of total sequences) candidate phyla, such as GN02, OP3, and PAUC34f (Figure 2c).
Below the phylum level, we observed little overlap between the kinds of bacteria that were specific to geographical location and those that were specific to host species. Bacteria associated with geographical locations were found in low abundance within animals, while those specific to a species were found in much greater abundance. This was extremely important for the natural products connection to symbiosis, as wherever we knew which bacteria produced ascidian natural products, they were among the most abundant (top-10 strains in terms of sequence abundance) in the animal (Figure 1; Supplementary Figure S2C). In other words, there was a strong connection between abundant symbionts and specific natural products.
Using the analyses employed, we were able to discern individual bacterial sequences responsible for location specificity (perhaps, environmentally derived sequences) from those that are host specific, which might be maintained in symbiosis with the hosts (Supplementary Table S6). Where the secondary metabolite producer was known in an ascidian species, this was among the major and species-specific strains found in that ascidian. For example, E. turbinata samples (Figure 3b, group 5) have two major, species-specific Gammaproteobacteria, one from the order Legionellanes (13–39% of sequenced reads, OTU 3550) and the other from an unclassified group (23–35% of sequenced reads, OTU 1589). The former is identical to Candidatus Endoecteinascidia frumentensis, which produces the potent anticancer metabolites, ecteinascidins within the ascidian E. turbinata (Rath et al., 2011). In another secondary metabolite-relevant example, L. patella from Fiji and Papua New Guinea (Figure 3b; group 1) differed from the other ascidians in the study by the high abundance of Prochloron spp. (58–80%). Prochloron spp. produce the family of cytotoxic cyclic peptides, cyanobactins, that are abundant, species-specific chemicals found in these ascidians (Schmidt et al., 2005). In two L. patella samples, FJ_06037 and PNG_11033, we were also able to identify a species-specific population of Alphaproteobacteria (OTU 932), which was the patellazole producer Ca. E. faulkneri. The presence of these known secondary metabolites was individually confirmed in each of these samples.
Even in samples where the secondary metabolite producers are unknown, host-specific bacterial sequences could be clearly discerned. For example, sequences that were species specific to California Didemnum sp. (see Figure 3b, group 3) include Gammaproteobacteria from the family Pseudomonadaceae and a Deltaproteobacteria from the genus Bdellovibrio.
L. badium microbiomes (Figure 3b, group 2) shared a major Deltaproteobacteria component from the order Sva0853 (16–64% of sequenced reads), an uncultivated order that was previously reported from seawater and coldwater sediments (Shi et al., 2011). The sequence itself was unique and represented an ascidian-specific OTU. Another group of Deltaproteobacteria in L. badium samples were the Desulfobacteria from the genus Nitrospina (Supplementary Figure S2; Supplementary Table S6). This is consistent with the recent report on the microbiome of ascidians from the Great Barrier Reef, where Nitrospina spp. were also found in high abundance in L. badium (Erwin et al., 2013). Nitrospina spp. are nitrite oxidizers and are proposed to have key roles in nitrogen fixing and cycling within the ascidian tunics (Erwin et al., 2013; Lucker et al., 2013). Another group that was slightly enriched in L. badium samples specifically in PNG_11062 (22%) is the Acidimicrobiales class of actinobacteria, a group that was previously described as a sponge-specific bacterial group (Simister et al., 2012). All the analyses for identifying the most abundant species-specific bacteria were further confirmed by Metastats analyses for the statistical identification of differentially abundance OTUs (data not shown).
By contrast, the location-specific component of ascidians represented sequences that are relatively common in the environment. Examples include OTU 3071, a Gammaproteobacterium from the order Chromatiales (average 0.16% in samples), and OTU 116, an Alteromonas sp. (average 0.10% in samples), both from California samples. In samples from Florida, unclassified Gammaproteobacteria (OTU 701, 0.17%), Vibrio spp. (OTU 766, 0.11%) and Oceanospirillales spp. (OTU 1931, 0.38%) are among the groups that are shared by the ascidians regardless of species.
Other notable bacteria found in sequenced samples
A subset of strains that might be of interest to the ascidian natural products community is described in this section. A nitrogen-fixing bacterium, Mesorhizobium sp., found in lower abundance in L. badium, was enriched in Cystodytes sp. samples (8–21%). This group has been reported from Cystodytes sp. before and was confirmed by BLAST analysis to the database sequences (Martinez-Garcia et al., 2007). Marine Mesorhizobium spp. have been shown to produce quorum-sensing molecules (Krick et al., 2007). The remainder of the Cystodytes sp. bacterial associates are distinct from those found in other ascidians. As an example, Cystodytes sp. samples contain Microbulbifer sp., a group known for their ability to degrade complex carbohydrates (Peng et al., 2006), at a relatively higher abundance (3–7% of sequences) as compared with other samples (0–0.3% of sequences).
The lowest diversity of sequences was found in Eudistoma sp. 004, with 92% (out of 13 886 sequences) comprising a species from the Endozoiconomonas family of the order Oceanospirillales (Supplementary Figure S2B). This group of bacteria has been reported in association with other marine organisms, including gorgonians, sea anemones and corals (Bayer et al., 2013; Correa et al., 2013; Lema et al., 2013). Interestingly, in most cases where they were present, they were also found to be dominant in the microbiome, leading to speculation about their possible symbiotic roles (Schuett et al., 2007).
Sequences from Florida Didemnum sp. TB3 were 22% Myxococcales (Supplementary Figure S2A). Myxobacteria have rarely been found in marine habitats (Reichenbach, 1999). Myxobacteria are prolific producers of natural products, and the first marine obligate halophilic myxobacteria-derived natural products were only reported very recently (Felder et al., 2013). Didemnum sp. TB3 also contained Flavobacteria from the genus Tenacibaculum (12%), a group that had been isolated from sponges and marine algae. Members of this group have also been reported as producers of hydroxamate siderophores (Fujita et al., 2013).
The microbiome of the solitary ascidian S. plicata was recently described from a Mediterranean sample (Erwin et al., 2013). We analyzed one sample of this species that we collected in California. Comparison of the composition at the phylum level of these samples showed that some of the major phyla such as Proteobacteria and Bacteriodetes are maintained, while some are not, perhaps owing to their ability to establish varied interactions upon introduction to new environments (Erwin et al., 2013). Our sample contained high levels of sequences assigned to chloroplasts, which could indicate an algal origin, while the Mediterranean samples harbored high levels of Planctomycetes.
Bioactivity profile of ascidian chemical extracts
We assayed the chemical extracts of the ascidians against laboratory strains of microorganisms to test whether there are distinct differences in the potency and toxicity of metabolites from the tropical regions compared with the temperate regions. We tested for activities against different microbes, including Staphylococcus aureus, Escherichia coli and Mycobacterium tuberculosis. We also tested for cytotoxicity against a human CEM-TART cell line and for antifungal activity against Candida albicans. There is a clear geographical distribution of biological activity and potency in these samples, which reflects the results of the secondary metabolite analysis performed as described below (Supplementary Figure S8). In these assays, the most potent extracts were those of tropical ascidians while the least potent are those of temperate ascidians. Importantly, the toxicity of these samples was not correlated to diversity of the underlying microbiome but was independent of this variable. This indicates that strains with more diverse microbiomes do not produce more toxic compounds and serves to further reinforce the role of single, talented bacterial symbionts in the production of toxins (see Discussion).
Chemistry of ascidians
PCA and hierarchical clustering of the LC/MS profiles showed that ascidians generally grouped by geographical location rather than by species (Figure 5). Three major clusters are formed comprising California, Florida and a combination of Papua New Guinea and Vanuatu in a single cluster. We hypothesized that this pattern was due to lipids in the extracts, which likely vary, because different temperatures would require different components to maintain membrane fluidity (Parrish, 2013). Alternatively, it might be due to other effects, such as available food sources, although this seemed less likely due to the large geographical sampling range in the tropical Pacific. Using the PCA biplot of ascidians and compounds (Supplementary Figure S5A), we performed an initial identification of the extract components that led to the geographical clustering of the ascidians. The chromatographic peaks were assigned using Lipid Maps (Fahy et al., 2007), and the resulting peaks assigned to lipids were then applied to PCA analysis. By using this putative lipid subset, we observed the same clustering by geographical location in the PCA (Supplementary Figure S5B). Conversely, we also subtracted the Lipid Maps-assigned peaks from the whole mzxml files and subjected the remaining data to PCA. Upon removing putative lipids from the analysis, the clustering according to geographical location was no longer observed. Instead, clustering was better correlated with animals species (see below).
Geographical clustering of ascidian samples due to lipid components. (a) PCA plot of ascidians in the first two dimensions showing clustering by geographical location (FL, Florida; PNG, Papua New Guinea; VN, Vanuatu; CA, California). (b) Example of how lipids were identified in this study. LC-MS/MS analysis and comparison of fragmentation patterns and accurate mass to lipids in METLIN database allowed assignment of specific fragments (colored arrows) to their complementary molecular components within lipid structures (colored boxes surrounding lipid molecule). Although these fragments do not absolutely confirm lipid identity, they provide high confidence that the compound falls within the lipid family as shown. See Supplementary Figures S5 and S6 for more details of lipid analysis.
A weakness of all MS-based approaches is that it is difficult to be sure that an ion has been accurately assigned. We therefore used further methods to determine whether ions preliminarily assigned as lipids did indeed truly represent lipids. We selected representative extracts from each PCA cluster and analyzed them using a validated metabolomics method, in which fragmentation of lipids (MS/MS profile) was compared with a large database of lipid fragment patterns. Through this analysis, we confirmed that the major components responsible for variance in the PCA were lipids. These included phospho-glycerolipids, glycerolipids and some straight chain fatty acids (Figure 5, Supplementary Figure S6).
We further analyzed the ascidian chemistry between samples within the same geographical location. We focused on Papua New Guinea samples where we have groups containing replicates of the same species and have characterized the natural products chemistry (Donia et al., 2011b). By PCA, we found that the samples cluster by host species. For example, L. patella samples (one from Fiji and two from Papua New Guinea) formed a separate cluster that was strongly driven by secondary metabolites. These metabolites include members of the cyanobactin family: patellins 1, 2, and 5 (Carroll et al., 1996); and the patellazoles (Corley et al., 1988; Zabriskie et al., 1988) (Figure 6, Supplementary Figure S7). Similarly, L. badium samples grouped together and in correlation with varamines (Molinski and Ireland, 1989) and some other unknown metabolites. When we included the Cystodytes sp. samples from Vanuatu in this analysis, these also strongly clustered together as expected, except for a single sample that was phylogenetically different than the others (Supplementary Figure S1). The grouping of Cystodytes samples was also caused by secondary metabolites, mostly pyridoacridine alkaloids (Marshall and Barrows, 2004), and the variations within the species were due to the abundance of different compound analogs. Specifically, VN_08002 and VN_08041 samples contained major amounts of cystodytins while VN_08019 mainly contained shermilamines in the extract (Cooray et al., 1988; Kobayashi et al., 1991).
Total ion chromatograms of organic extracts of selected ascidians that are known for secondary metabolite production showing the abundance and host specificity of secondary metabolites in ascidian extracts. The secondary metabolites represent the major compounds in the extracts, and, as shown in Supplementary Figure S7, are highly species specific.
In cases where they could be identified, secondary metabolites were also noticeably major components of extracts of other ascidians from Florida, such as the eudistomins (Kobayashi et al., 1984) from Eudistoma sp. TB1. The extract of L. bistratum also contained major metabolites, including the bistramides and the bistratamides group of compounds (Degnan et al., 1989; Biard et al., 1994; Perez and Faulkner, 2003). However, in most samples, especially those from California, the majority of the components could not be assigned from databases, such as Antimarin and the Dictionary of Natural Products.
Discussion
We set out to determine whether there are any correlations between ascidian bacterial communities and the presence of abundant, bioactive secondary metabolites. By examining phylogenetically diverse ascidians across diverse locations and a large latitudinal gradient, we hoped to differentiate factors related to location and habitat from those related to metabolism. At the level of common indices of overall bacterial diversity, ascidian microbiomes were virtually identically diverse across all sample types and habitats. With a few exceptions, ascidians generally maintain rich and diverse microbiomes in several widely different locations and environments and thus provide untapped sources of microbial diversity. Perhaps owing to their similar lifestyle, this diversity is comparable to that of sponges (Schmitt et al., 2012).
Our results show that chemical diversity in ascidians is not reflected at the microbiome diversity level but rather in specific interactions with talented bacterial producers. We propose that, in the tropical regions where there is more pressure for defensive compounds, selection leads to symbiosis with bacteria that produce defensive metabolites, leading to a higher fitness for the associated organisms. In this model, the host species-specific bacteria produce the species-specific secondary metabolites isolated from ascidians. The bacterial producers of ascidian compounds are only known in a few cases, including in several samples described in this study. The sources of by far most ascidian metabolites remain unknown. From this study, it is clear that ascidians do not have universal solutions to this problem and that ascidian species have widely different microbiomes, in which one or two talented producers may be responsible for producing the resulting metabolites.
Ascidian microbiomes were stable over time and space. The majority of these microbes compose the species-specific microbiome, which consisted of more abundant (‘top-10’) species of bacteria in each sample. These species-specific microbiomes often contained relatively rare types of bacteria. Low-abundance bacteria on the other hand tended to be relatively widespread species, which were strongly correlated with the geography of the samples. Such species may be associated with changes in seawater or environment, and their specific association with ascidians is uncertain.
Several studies have previously examined some of the species of ascidians included in this work, further extending that analysis to several different locations and water temperature regimes. Mediterranean S. plicata microbiomes were recently examined in detail (Erwin et al., 2013) and revealed key similarities with the California sample studied here. L. badium from the Great Barrier Reef (Erwin et al., 2013) had a very similar microbiome to samples examined here from Papua New Guinea. The L. patella microbiome has been examined by both whole genome sequencing methods in our laboatory using samples from Palau, Solomon Islands, Fiji and Papua New Guinea (Donia et al., 2011a, 2011b) and by 16S-based methods using Great Barrier Reef samples (Behrendt et al., 2012). Here, we extended these analyses to further 16S-based methods across additional samples, further confirming the species specificity previously reported. Finally, E. turbinata has been examined by whole genome sequencing (Rath et al., 2011) and by 16S-based analysis (Moss et al., 2003; Perez-Matos et al., 2007), with deeper sequences obtained here. The comparison here of multiple species across time and space reveals common elements of diversity in ascidian microbiomes.
This study is the first to compare ascidian metabolomes and microbiomes across different species and geographical locations. As with the microbiomes, the metabolomes had location-specific and species-specific components. The metabolites responsible for most of the observed variance in PCA analysis were shown to be lipids. We propose that this difference may be due to the different temperatures at the varied sampling sites (∼10 °C to ∼30 °C across the latitudinal gradient) (Parrish, 2013). Once the influence of the lipids were removed from the analysis, or focusing on samples from single regions, a strong species-specific component was revealed that was due largely to secondary metabolism (Figure 6). Thus both the microbiome and the major secondary metabolites were strongly species specific. In the relatively limited sample set studied, the tropical ascidians were highly bioactive, exhibiting potent toxicity to diverse cell types, whereas subtropical (Florida) samples were less toxic, and temperate samples nearly lacked toxicity. The compounds responsible for activity in tropical samples were mostly known compounds, exceptionally potent marine natural products such as patellazoles and pyridoacridines that may serve to defend their hosts. Within the subset of L. patella, secondary metabolism is highly correlated to phylogeny of the host even within this single species (Kwan et al., 2014). This analysis extends this observation across multiple species, showing that secondary metabolism is strongly related to ascidian phylogeny, and the apparently random distribution of secondary metabolites within a single species previously observed is likely due to lack of appropriate measures of phylogenetic divergence.
In this study, we provide an overview of the key players in the ecology of chemical defense in ascidians: bacteria and secondary metabolites. Bacterial diversity or abundance is not a marker for chemical diversity or the potency of secondary metabolites in ascidians. Instead, ascidians harbor species-specific microbiomes and species-specific secondary metabolites (often, defensive compounds). Where the producer of compounds is known, these producers represent one of the top-10 strains found in ascidian microbiomes. This implies that, in the search for natural products and their bacterial producers, efforts should be primarily focused on the most abundant bacteria in the microbiota, such that only perhaps 10 strains may be examined to find a producer. The known bacterial producers of ascidian secondary metabolites are phylogenetically diverse, and they are ascidian species-specific and ancient in their distribution (Kwan and Schmidt, 2013). It is remarkable that many different associations with different bacteria have led to defensive chemicals in ascidians, implying a strong selection for this property and the independent origin of defensive associations in different ascidian species.
Accession codes
Accessions
GenBank/EMBL/DDBJ
Sequence Read Archive
References
Angiuoli SV, Matalka M, Gussman A, Galens K, Vangala M, Riley DR et al. (2011). CloVR: a virtual machine for automated and portable sequence analysis from the desktop using cloud computing. BMC Bioinformatics 12: 356.
Bayer T, Neave MJ, Alsheikh-Hussain A, Aranda M, Yum LK, Mincer T et al. (2013). The microbiome of the Red Sea coral Stylophora pistillata is dominated by tissue-associated Endozoicomonas bacteria. Appl Environ Microbiol 79: 4759–4762.
Behrendt L, Larkum AW, Trampe E, Norman A, Sorensen SJ, Kuhl M . (2012). Microbial diversity of biofilm communities in microniches associated with the didemnid ascidian Lissoclinum patella. ISME J 6: 1222–1237.
Biard JF, Roussakis C, Kornprobst JM, Gouiffes-Barbin D, Verbist JF, Cotelle P et al. (1994). Bistramides A, B, C, D, and K: a new class of bioactive cyclic polyethers from Lissoclinum bistratum. J Nat Prod 57: 1336–1345.
Caporaso JG, Bittinger K, Bushman FD, DeSantis TZ, Andersen GL, Knight R . (2010a). PyNAST: a flexible tool for aligning sequences to a template alignment. Bioinformatics 26: 266–267.
Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK et al. (2010b). QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7: 335–336.
Carroll AR, Coll JC, Bourne DJ, MacLeod JK, Zabriskie TM . (1996). Patellins 1-6 and trunkamide A: novel cyclic hexa-, hepta- and octa-peptides from colonial ascidians, Lissoclinum sp. Aust J Chem 49: 659–667.
Castresana J . (2000). Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol 17: 540–552.
Chapman MG, Underwood AJ . (1999). Ecological patterns in multivariate assemblages: Information and interpretation of negative values in ANOSIM tests. Mar Ecol Prog Ser 180: 257–265.
Clarke KR . (1993). Non-parametric multivariate analyses of changes in community structure. Aust J Ecol 18: 117–143.
Cooray NM, Scheuer PJ, Parkanyi L, Clardy J . (1988). Shermilamine A: a pentacyclic alkaloid from a tunicate. J Org Chem 53: 4619–4620.
Corley DG, Moore RE, Paul VJ . (1988). Patellazole B: a novel cytotoxic thiazole-containing macrolide from the marine tunicate Lissoclinum patella. J Am Chem Soc 110: 7920–7922.
Correa H, Haltli B, Duque C, Kerr R . (2013). Bacterial communities of the gorgonian octocoral Pseudopterogorgia elisabethae. Microb Ecol 66: 972–985.
Degnan BM, Hawkins CJ, Lavin MF, McCaffrey EJ, Parry DL, Watters DJ . (1989). Novel cytotoxic compounds from the ascidian Lissoclinum bistratum. J Med Chem 32: 1354–1359.
DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K et al. (2006). Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol 72: 5069–5072.
Dishaw LJ, Flores-Torres J, Lax S, Gemayel K, Leigh B, Melillo D et al. (2014). The gut of geographically disparate Ciona intestinalis harbors a core microbiota. PLoS One 9: e93386.
Donia MS, Hathaway BJ, Sudek S, Haygood MG, Rosovitz MJ, Ravel J et al. (2006). Natural combinatorial peptide libraries in cyanobacterial symbionts of marine ascidians. Nat Chem Biol 2: 729–735.
Donia MS, Ravel J, Schmidt EW . (2008). A global assembly line for cyanobactins. Nat Chem Biol 4: 341–343.
Donia MS, Fricke WF, Partensky F, Cox J, Elshahawi SI, White JR et al. (2011a). Complex microbiome underlying secondary and primary metabolism in the tunicate-Prochloron symbiosis. Proc Natl Acad Sci USA 108: E1423–E1432.
Donia MS, Fricke WF, Ravel J, Schmidt EW . (2011b). Variation in tropical reef symbiont metagenomes defined by secondary metabolism. PLoS One 6: e17897.
Donia MS, Ruffner DE, Cao S, Schmidt EW . (2011c). Accessing the hidden majority of marine natural products through metagenomics. Chembiochem 12: 1230–1236.
Edgar RC . (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32: 1792–1797.
Edgar RC . (2010). Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26: 2460–2461.
Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R . (2011). UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27: 2194–2200.
Erwin PM, Pineda MC, Webster N, Turon X, Lopez-Legentil S . (2013). Down under the tunic: bacterial biodiversity hotspots and widespread ammonia-oxidizing archaea in coral reef ascidians. ISME J 8: 575–588.
Erwin PM, Pineda MC, Webster N, Turon X, Lopez-Legentil S . (2013). Small core communities and high variability in bacteria associated with the introduced ascidian Styela plicata. Symbiosis 59: 35–46.
Fahy E, Sud M, Cotter D, Subramaniam S . (2007). LIPID MAPS online tools for lipid research. Nucleic Acids Res 35: W606–W612.
Felder S, Dreisigacker S, Kehraus S, Neu E, Bierbaum G, Wright PR et al. (2013). Salimabromide: unexpected chemistry from the obligate marine myxobacterium Enhygromxya salina. Chemistry 19: 9319–9324.
Fujita MJ, Nakano K, Sakai R . (2013). Bisucaberin B, a linear hydroxamate class siderophore from the marine bacterium Tenacibaculum mesophilum. Molecules 18: 3917–3926.
Hirose E, Fukuda T . (2006). Vertical transmission of photosymbionts in the colonial ascidian Didemnum molle: the larval tunic prevents symbionts from attaching to the anterior part of larvae. Zoolog Sci 23: 669–674.
Hirose E, Hirabayashi S, Hori K, Kasai F, Watanabe MM . (2006). UV protection in the photosymbiotic ascidian Didemnum molle inhabiting different depths. Zoolog Sci 23: 57–63.
Hirose E, Neilan BA, Schmidt EW, Murakami A . (2009). Enigmatic life and evolution of Prochloron and related cyanobacteria inhabiting colonial ascidians. Handbook on Cyanobacteria. Nova Science Publishers Inc.: Hauppauge, NY, USA, pp 161–189.
Hou Y, Braun DR, Michel CR, Klassen JL, Adnani N, Wyche TP et al. (2012). Microbial strain prioritization using metabolomics tools for the discovery of natural products. Anal Chem 84: 4277–4283.
Joullie MM, Leonard MS, Portonovo P, Liang B, Ding X, La Clair JJ . (2003). Chemical defense in ascidians of the didemnidae family. Bioconjug Chem 14: 30–37.
Kobayashi J, Harbour GC, Gilmore J, Rinehart KL . (1984). Eudistomins A, D, G, H, I, J, M, N, O, P, and Q, bromo, hydroxy, pyrrolyl and iminoazepino.beta.-carbolines from the antiviral Caribbean tunicate Eudistoma olivaceum. J Am Chem Soc 106: 1526–1528.
Kobayashi J, Tsuda M, Tanabe A, Ishibashi M, Cheng JF, Yamamura S et al. (1991). Cystodytins D-I, new cytotoxic tetracyclic aromatic alkaloids from the Okinawan marine tunicate Cystodytes dellechiajei. J Nat Prod 54: 1634–1638.
Kohl M, Wiese S, Warscheid B . (2011). Cytoscape: software for visualization and analysis of biological networks. Methods Mol Biol 696: 291–303.
Kojima A, Hirose E . (2010). Transfer of prokaryotic algal symbionts from a tropical ascidian (Lissoclinum punctatum) colony to its larvae. Zoolog Sci 27: 124–127.
Krick A, Kehraus S, Eberl L, Riedel K, Anke H, Kaesler I et al. (2007). A marine Mesorhizobium sp. produces structurally novel long-chain N-acyl-L-homoserine lactones. Appl Environ Microbiol 73: 3587–3594.
Kwan JC, Donia MS, Han AW, Hirose E, Haygood MG, Schmidt EW . (2012). Genome streamlining and chemical defense in a coral reef symbiosis. Proc Natl Acad Sci USA 109: 20655–20660.
Kwan JC, Schmidt EW . (2013). Bacterial endosymbiosis in a chordate host: long-term co-evolution and conservation of secondary metabolism. PLoS One 8: e80822.
Kwan JC, Tianero MDB, Donia MS, Wyche TP, Bugni TS, Schmidt EW . (2014). Host control of symbiont natural product chemistry in cryptic populations of the tunicate Lissoclinum patella. PLoS One 9: e95850.
Lema KA, Willis BL, Bourne DG . (2013). Amplicon pyrosequencing reveals spatial and temporal consistency in diazotroph assemblages of the Acropora millepora microbiome. Environ Microbiol e-pub ahead of print 24 December 2013 doi:10.1111/1462-2920.12366.
Letunic I, Bork P . (2011). Interactive Tree Of Life v2: online annotation and display of phylogenetic trees made easy. Nucleic Acids Res 39: W475–W478.
Lozupone C, Lladser ME, Knights D, Stombaugh J, Knight R . (2011). UniFrac: an effective distance metric for microbial community comparison. ISME J 5: 169–172.
Lucker S, Nowka B, Rattei T, Spieck E, Daims H . (2013). The genome of nitrospina gracilis illuminates the metabolism and evolution of the major marine nitrite oxidizer. Front Microbiol 4: 27.
Marshall KM, Barrows LR . (2004). Biological activities of pyridoacridines. Nat Prod Rep 21: 731–751.
Martinez-Garcia M, Diaz-Valdes M, Wanner G, Ramos-Espla A, Anton J . (2007). Microbial community associated with the colonial ascidian Cystodytes dellechiajei. Environ Microbiol 9: 521–534.
McMurdie PJ, Holmes S . (2013). phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One 8: e61217.
Molinski TF, Ireland CM . (1989). Varamines A and B, new cytotoxic thioalkaloids from Lissoclinum vareau. J Org Chem 54: 4256–4259.
Monniot C, Monniot F, Laboute P . (1991) Coral Reef Ascidians of New Caledonia. Editions de 'ORSTOM: Paris, France.
Moss C, Green DH, Perez B, Velasco A, Henriquez R, McKenzie JD . (2003). Intracellular bacteria associated with the ascidian Ecteinascidia turbinata: Phylogenetic and in situ hybridisation analysis. Mar Biol 143: 99–110.
Oksanen J, Blanchet FG, Kindt R, Legendre P, Minchin PR, O' Hara RB et al. (2013). vegan: Community Ecology Package, R Package version 2.0-8 edn.
Pallmann P, Schaarschmidt F, Hothorn LA, Fischer C, Nacke H, Priesnitz KU et al. (2012). Assessing group differences in biodiversity by simultaneously testing a user-defined selection of diversity indices. Mol Ecol Resour 12: 1068–1078.
Parrish C . (2013). Lipids in marine ecosystems. ISRN Oceanogr 2013: article ID 604045.
Paul VJ, Lindquist N, Fenical W . (1990). Chemical defenses of the tropical ascidian Atapozoa sp. and its nudibranch predators Nembrotha spp. Mar Ecol Prog Ser 59: 109–118.
Peng X, Adachi K, Chen C, Kasai H, Kanoh K, Shizuri Y et al. (2006). Discovery of a marine bacterium producing 4-hydroxybenzoate and its alkyl esters, parabens. Appl Environ Microbiol 72: 5556–5561.
Perez LJ, Faulkner DJ . (2003). Bistratamides E-J, modified cyclic hexapeptides from the Philippines ascidian Lissoclinum bistratum. J Nat Prod 66: 247–250.
Perez-Matos AE, Rosado W, Govind NS . (2007). Bacterial diversity associated with the Caribbean tunicate Ecteinascidia turbinata. Antonie Van Leeuwenhoek 92: 155–164.
Pluskal T, Castillo S, Villar-Briones A, Oresic M . (2010). MZmine 2: modular framework for processing, visualizing, and analyzing mass spectrometry-based molecular profile data. BMC Bioinformatics 11: 395.
Price MN, Dehal PS, Arkin AP . (2010). FastTree 2—approximately maximum-likelihood trees for large alignments. PLoS One 5: e9490.
Rath CM, Janto B, Earl J, Ahmed A, Hu FZ, Hiller L et al. (2011). Meta-omic characterization of the marine invertebrate microbial consortium that produces the chemotherapeutic natural product ET-743. ACS Chem Biol 6: 1244–1256.
Reichenbach H . (1999). The ecology of the myxobacteria. Environ Microbiol 1: 15–21.
Riesenfeld CS, Murray AE, Baker BJ . (2008). Characterization of the microbial community and polyketide biosynthetic potential in the palmerolide-producing tunicate Synoicum adareanum. J Nat Prod 71: 1812–1818.
Schmidt EW, Nelson JT, Rasko DA, Sudek S, Eisen JA, Haygood MG et al. (2005). Patellamide A and C biosynthesis by a microcin-like pathway in Prochloron didemni, the cyanobacterial symbiont of Lissoclinum patella. Proc Natl Acad Sci USA 102: 7315–7320.
Schmidt EW, Donia MS . (2009). Chapter 23. Cyanobactin ribosomally synthesized peptides—a case of deep metagenome mining. Methods Enzymol 458: 575–596.
Schmidt EW, Donia MS . (2010). Life in cellulose houses: symbiotic bacterial biosynthesis of ascidian drugs and drug leads. Curr Opin Biotechnol 21: 827–833.
Schmidt EW, Donia MS, McIntosh JA, Fricke WF, Ravel J . (2012). Origin and variation of tunicate secondary metabolites. J Nat Prod 75: 295–304.
Schmitt S, Tsai P, Bell J, Fromont J, Ilan M, Lindquist N et al. (2012). Assessing the complex sponge microbiota: core, variable and species-specific bacterial communities in marine sponges. ISME J 6: 564–576.
Schuett C, Doepke H, Grathoff A, Gedde M . (2007). Bacterial aggregates in the tentacles of the sea anemone Metridium senile. Helgol Mar Res 61: 211–216.
Shenkar N, Swalla BJ . (2011). Global diversity of Ascidiacea. PLoS One 6: e20657.
Shi Y, Tyson GW, Eppley JM, DeLong EF . (2011). Integrated metatranscriptomic and metagenomic analyses of stratified microbial assemblages in the open ocean. ISME J 5: 999–1013.
Simister RL, Deines P, Botte ES, Webster NS, Taylor MW . (2012). Sponge-specific clusters revisited: a comprehensive phylogeny of sponge-associated microorganisms. Environ Microbiol 14: 517–524.
Sokolov EP . (2000). An improved method for DNA isolation from mucopolysaccharide-rich molluscan tissues. J Mollusc Stud 66: 573–575.
R Core Team. (2013) R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing: Vienna, Austria.
Wang Q, Garrity GM, Tiedje JM, Cole JR . (2007). Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol 73: 5261–5267.
Xu Y, Kersten RD, Nam SJ, Lu L, Al-Suwailem AM, Zheng H et al. (2012). Bacterial biosynthesis and maturation of the didemnin anti-cancer agents. J Am Chem Soc 134: 8625–8632.
Yokobori S, Kurabayashi A, Neilan BA, Maruyama T, Hirose E . (2006). Multiple origins of the ascidian-Prochloron symbiosis: molecular phylogeny of photosymbiotic and non-symbiotic colonial ascidians inferred from 18S rDNA sequences. Mol Phylogenet Evol 40: 8–19.
Zabriskie TM, Mayne CL, Ireland CM . (1988). Patellazole C: A novel cytotoxic macrolide from Lissoclinum patella. J Am Chem Soc 110: 7919–7920.
Acknowledgements
We thank James Cox and John Alan Maschek of the University of Utah Metabolomics core facility for assistance and help on the LC-MS/MS analysis. Appropriate permits were obtained for all collections reported in this manuscript. We are grateful to the staff of USC Wrigley Institute and, in particular, USC undergraduate Mariah Gill for aid with obtaining California samples. We thank Chris Ireland and Mary Kay Harper for the L. voeltzkawi sample that was obtained through the PNG-ICBG program and the Vanuatu samples that were collected through the NCDDG program. This investigation was supported by the University of Utah Study Design and Biostatistics Center, with funding in part from the National Center for Research Resources and the National Center for Advancing Translational Sciences, National Institutes of Health, through Grant 8UL1TR000105 (formerly UL1RR025764). This research was funded by NIH GM092009 and GM107557.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on The ISME Journal website
Supplementary information
Rights and permissions
About this article
Cite this article
Tianero, M., Kwan, J., Wyche, T. et al. Species specificity of symbiosis and secondary metabolism in ascidians. ISME J 9, 615–628 (2015). https://doi.org/10.1038/ismej.2014.152
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ismej.2014.152
This article is cited by
-
Prokaryotic symbiont communities in three ascidian species introduced in both Ireland and New Zealand
Environmental Science and Pollution Research (2023)
-
Genomic insights into symbiosis and host adaptation of an ascidian-associated bacterium Bacillus aryabhattai MCCB 387
Symbiosis (2022)
-
The bacterial composition associated with Atriolum robustum, a common ascidian from Xisha coral reef, China
Symbiosis (2021)
-
Existence in cellulose shelters: industrial and pharmaceutical leads of symbiotic actinobacteria from ascidian Phallusia nigra, Andaman Islands
World Journal of Microbiology and Biotechnology (2021)
-
Taxonomic Composition and Biological Activity of Bacterial Communities Associated with Marine Ascidians from Andaman Islands, India
Applied Biochemistry and Biotechnology (2021)