Abstract
The biosynthesis of non-ribosomal peptide and polyketide natural products is facilitated by multimodular enzymes that contain domains responsible for the sequential condensation of amino and carboxylic subunits. These conserved domains provide molecular targets for the discovery of natural products from microbial metagenomes. This study demonstrates the application of tag-encoded FLX amplicon pyrosequencing (TEFAP) targeting non-ribosomal peptide synthetase (NRPS) and polyketide synthase (PKS) genes as a method for determining the identity and diversity of natural product biosynthesis genes. To validate this approach, we assessed the diversity of NRPS and PKS genes within the microbiomes of six Australian marine sponge species using both TEFAP and metagenomic whole-genome shotgun sequencing approaches. The TEFAP approach identified 100 novel ketosynthase (KS) domain sequences and 400 novel condensation domain sequences within the microbiomes of the six sponges. The diversity of KS domains within the microbiome of a single sponge species Scopalina sp. exceeded that of any previously surveyed marine sponge. Furthermore, this study represented the first to target the condensation domain from NRPS biosynthesis and resulted in the identification of a novel condensation domain lineage. This study highlights the untapped potential of Australian marine sponges for the isolation of novel bioactive natural products. Furthermore, this study demonstrates that TEFAP approaches can be applied to functional genes, involved in natural product biosynthesis, as a tool to aid natural product discovery. It is envisaged that this approach will be used across multiple environments, offering an insight into the biological processes that influence the production of secondary metabolites.
Similar content being viewed by others
Introduction
Non-ribosomal peptides (NRPs) and polyketides (PKs) are the cornerstones of many modern pharmaceuticals, and provide a molecular and chemical source for the isolation of novel bioactive compounds. The last 30 years have seen the majority of these compounds derived from microorganisms that were isolated from the environment and cultured within the laboratory (Newman and Cragg, 2012). Although advances in culturing methodology are enabling the isolation of previously recalcitrant microbes (Ferrari et al., 2008), culturable organisms still represent only a small proportion of the total microbial diversity in many environments (Keller and Zengler, 2004; Donachie et al., 2007). The uncertainty regarding the depth of metabolic diversity that exists within a given sample remains.
Marine sponges represent a significant resource for the isolation of novel bioactive compounds (Fusetani and Matsunaga, 1993; Blunt et al., 2006; Thomas et al., 2010). The majority of these compounds exhibit structural features indicative of bacterial biosynthetic routes and microorganisms have been isolated that are capable of producing sponge-associated metabolites (Boot et al., 2006). Previous molecular studies, utilising vector-dependent approaches, identified a lack of functional diversity among some sponge species due to the dominance of sponge-specific polyketide synthases (PKSs) (Piel et al., 2004b; Schirmer et al., 2005; Fieseler et al., 2007; Hochmuth et al., 2010). This has since led to the development of assays that amplify specific ketosynthase (KS) groups involved in the biosynthesis of complex secondary metabolites (Piel et al., 2004a; Fisch et al., 2009). However, these highly specific approaches are only applicable for the identification of biosynthetic pathways when the product is known. The feasibility of using high-throughput sequencing methods to determine the true metabolic diversity of KS domains from a single marine sponge has been previously reported (Trindade-Silva et al., 2012). In contrast, those studies targeting non-ribosomal peptide synthetases (NRPS) from marine sponge microbiomes, limited to the use of vector-dependent approaches targeting the adenylation (A) domain, revealed minimal biosynthetic diversity (Kennedy et al., 2008; Pimentel-Elardo et al., 2012).
The biosynthesis of NRPs and PKs is facilitated by the action of a series of multimodular enzymes, with each module containing a core set of three domains required for the activation and condensation of a single amino or carboxylic subunit. The KS and condensation (C) domains, that catalyse the condensation of two activated subunits, contain motifs that are conserved at an amino-acid level (Stachelhaus et al., 1998; Walsh and Fischbach, 2010). Between these motifs, variation is observed, typically within catalytic centres, giving rise to domains with functional specificity, reflecting the nature of the subunits being condensed (Moffitt and Neilan, 2003; Rausch et al., 2007), the arrangement of other domains within the module and the expected product (Moffitt and Neilan, 2003; Piel et al., 2004a; Schmitt et al., 2008). The conserved nature for part of these domains ensures their suitability as targets for amplification using the PCR. Combining PCR with tag-encoded FLX amplicon pyrosequencing (TEFAP) (Sun et al., 2011) provides a possible means for evaluating the composition and diversity of NRPS and PKS genes from microbiomes. TEFAP generates thousands of reads per sample with sufficient read length (∼400 bp) and allows for phylogenetic discrimination amongst both taxonomic markers and functional genes, for example, nifH and dmdA. By multiplexing large numbers of environmental samples, the cost and time involved in evaluating the composition and diversity of a single target within an environment is drastically reduced.
The strength of targeting these domains is that they describe the diversity of gene clusters encoding bioactive molecules from organisms that are either difficult to isolate or produced by ‘rare biosphere’ representatives, as defined by their occurrence at very low abundances (Piel et al., 2004a; Sogin et al., 2006; Fisch et al., 2009). Traditional methods, involving exhaustive extractions of sponge, do not reveal the chemical diversity produced by these rare biosphere representatives, or pathways that are inactive within the native host (Chiang et al., 2010). Harnessing this potentially novel chemistry is, therefore, dependent on the ability of molecular approaches to identify where this diversity exists (Fieseler et al., 2007; Hochmuth et al., 2010; Trindade-Silva et al., 2012), to provide access to the genetic basis of biosynthesis (Piel et al., 2004a; Fisch et al., 2009; Banik and Brady, 2010; Brady et al., 2010) and ultimately to facilitate production of these molecules through heterologous expression within a suitable host (Fu et al., 2008; Craig et al., 2010).
In this study, we aim to highlight the untapped potential of Australian marine sponges by evaluating the diversity of NRP and PK biosynthesis genes using a deep-sequencing TEFAP approach. The limitations and advantages of the TEFAP method, in comparison to a non-targeted metagenomic whole-genome shotgun sequencing (mWGS) approach are determined by comparing the two independent outcomes. In addition, we provide insight into how these data sets could be applied for identifying and exploiting environments containing rich biosynthetic potential. Hence, this study demonstrates the suitability of a TEFAP-based approach for evaluating and improving access to the genetic basis for natural product biosynthesis within marine sponges and other environments with untapped genomic diversity.
Materials and methods
Degenerate PCR and TEFAP
DNA derived from microbial enrichments of six Australian marine sponges was obtained in triplicate as previously described (Fan et al., 2012). Extracted DNA from three replicates for each marine sponge species were pooled in equimolar amounts and screened using two sets of degenerate primers. KS sequences were amplified using the DKF/DKR (see Supplementary Information Table S1) primer pair as previously described (Moffitt and Neilan, 2003). C domain sequences from NRPS modules were amplified using the primers CnDmF and DCCR (see Supplementary Information Table S1). To facilitate subsequent incorporation of sample-specific barcodes and sequencing primer sites, amplification of each positive sample was repeated using the corresponding forward primer containing a 5′ T7 promoter sequence and the corresponding reverse primer containing a 5′ M13R sequence (see Supplementary Information Table S1). Amplification was performed for 35 rounds of thermal cycling at an annealing temperature of 50 °C for KS domains and 45 °C for C domains. Following PCR amplification products were gel extracted using the ZymoClean Gel Extraction Kit (Zymo Research, Irvine, CA, USA). For each positive sample 10 ng of purified PCR product was used as a template in a second PCR using the same cycling conditions. In this PCR a single universal reverse primer (454R-M13R) and sample-specific forward primer (454F-T7Prom) were used to incorporate priming sites of the sequencing primer and sample-specific barcodes (see Supplementary Tables S1 and S2). Samples were combined into two separate pools corresponding to each gene target. Library preparation and unidirectional amplicon sequencing was performed at the Clive and Vera Ramaciotti Centre for Gene Function Analysis (Sydney, Australia) using the 454 FLX Titanium platform.
Identification of NRPS and PKS from mWGS data
Metagenomic DNA for the microbial communities was isolated for each replicate of the six sponges and sequenced separately on a 454 FLX pyrosequencer (Roche, Branford, CT, USA) (Fan et al., 2012). Samples received between 360 000 and 1300 000 shotgun reads which were subsequently separately assembled using the Newbler software (Roche). Amino-acid sequences, translated from metagenomic contigs, for six sponges were obtained as previously described (Fan et al., 2012). Sequences containing KS and C domains were recovered using the HMMER algorithm (Finn et al., 2011) by searching the PFAM profiles (Finn et al., 2010) PF00109 and PF00668, respectively. Hits that obtained a bit score >25 were retained. For each protein sequence recovered, the corresponding nucleotide sequence was retained for further analysis.
Sequence processing
TEFAP reads were subjected to initial pre-processing that included noise reduction using the shhh.flows algorithm (Schloss et al., 2009), removal of sequences containing ambiguous bases and long (>8) homopolymers, multiplex-barcode dependent binning of sequences and removal of primer sequences.
Reference nucleotide and amino-acid alignments were generated using MUSCLE (Edgar, 2004), using reference sequences obtained from the Genbank database (Accessed: 23 January 2012) (Benson et al., 1997). Reference sequences were selected on the basis of either belonging to known, characterised biosynthetic pathways or due to their previously established phylogenetic distribution (Moffitt and Neilan, 2003; Roongsawang et al., 2005; Fieseler et al., 2007; Rausch et al., 2007). As a result, the C domain alignment contained 162 reference sequences while the KS domain alignment had 190 reference sequences.
Pre-processed TEFAP reads were screened against the respective nucleotide alignment to remove non-target sequences that were amplified as a result of the degenerate PCR conditions. Nucleotide sequences, derived from PFAM-dependent screening of the mWGS data sets, were also screened using this same approach. Following screening, the TEFAP reads and mWGS (open reading frames) were combined. Previous phylogenetic analyses of KS and C domains have indicated that these domains typically exhibit at least 5% amino-acid dissimilarity (Moffitt and Neilan, 2003; Roongsawang et al., 2005; Rausch et al., 2007). Previous TEFAP methods have reflected this amino-acid dissimilarity by clustering nucleotides at a 90% nucleotide similarity cutoff (Varaljay et al., 2010). Therefore, in this study, clustering of sequences into operational taxonomic units (OTUs) (Schloss and Westcott, 2011), rarefaction analyses, and generation of diversity indices was performed using the average neighbour method (Schloss et al., 2009) as implemented in Mothur version 1.23.1 at a 0.10 distance threshold.
Taxonomic and functional classification
To determine whether the two methods accessed comparable genetic diversities, a qualitative assessment of the approximate taxonomic identity of sequences from both methods was made. TEFAP OTUs and mWGS open reading frames were assigned to taxa using the last common ancestor algorithm incorporated in the MEGAN 4.70.4 software package (Huson et al., 2011). Individual sequences were assigned to a taxon where at least 10% of hits with a bit score >35 were in agreement. A direct comparison was also made by matching TEFAP OTUs with unprocessed mWGS sequences using the BLASTn algorithm (Altschul et al., 1997). A successful match was identified where at least 90% nucleotide identity was observed across at least 100 bp. The GC content of TEFAP reads and unprocessed mWGS was determined using the GEECEE algorithm in the EMBOSS package (Rice et al., 2000).
TEFAP and mWGS OTU representative sequences were individually scrutinised for correct translation into amino-acid sequences. Derived amino-acid sequences were aligned against the amino-acid reference alignment by MUSCLE (Edgar, 2004). Phylogenetic inference of amino-acid sequences was made using PhyML v3.0 (Guindon et al., 2005).
Results
Identification of novel NRPS and PKS biosynthetic genes by TEFAP
Using degenerate primers, PKSs (KS domains) were detected in Cymbastela concentrica, Cymbastela coralliophila, Sylissa sp., Scopalina sp. and Rhopaloeides odorabile, while NRPSs (C domains) were detected in Scopalina sp., C. concentrica and C. coralliophila. NRP and PK biosynthetic pathways were not detected in the microbiome of the sponge Tedania anhelens. Following processing through the pipeline described, approximately two-thirds of the sequences generated were discarded due to length and specificity requirements (see Supplementary Figure S1). In total, 1097 KS and 4469 C domain sequences of at least 240 bp in length were retained.
A search of PFAM sequence profiles recovered 2012 KS and 133 C domain sequences from the six mWGS data sets. The corresponding nucleic acid sequences were obtained with the length of KS domain sequences ranging between 87 and 1885 bp and the length of C domain sequences ranging between 79 and 792 bp. These nucleic acid sequences were subjected to the same pipeline that was applied to the TEFAP reads and 847 KS and 14 C domain sequences were retained (see Supplementary Figure S1). The low retention rate reflected the fragmented nature and short read length of the mWGS reads, resulting in the removal of partial KS domain and C domain sequences that did not contain the targeted domain region. Manual inspection, using the BLASTx algorithm, revealed many of the 847 KS sequences corresponded to ketoacylsynthase (I/II) sequences that, while belonging to the KS superfamily (Moffitt and Neilan, 2003), relate to aspects of primary metabolism. Following removal of these sequences, 114 KS sequences remained (Table 1). Processed TEFAP and mWGS sequences were combined and clustered into OTUs at a 0.10 distance threshold resulting in the generation of 133 KS domain OTUs and 396 C domain OTUs (Table 1). To assess whether any reads corresponded to previously characterised biosynthetic pathways, dereplication of TEFAP and mWGS OTUs was performed by comparison against the Genbank database (Benson et al., 1997) using the BLASTx algorithm (Altschul et al., 1997). Only one C domain and 10 KS TEFAP OTUs exhibited significant (>90%) translated amino-acid identity to previously identified NRPS and PKS protein sequences, respectively.
Comparison of TEFAP and mWGS approaches for the recovery of KS and C domain diversity
TEFAP reads were in excess of mWGS sequences in each sample for both gene targets. As a consequence, the number of OTUs observed by the TEFAP approach exceeded that observed by the mWGS approach (Table 1; Supplementary Figure S2), indicating a more comprehensive evaluation of the diversity of these secondary metabolite biosynthesis genes. The only exception was the sponge R. odorabile, in which more OTUs were observed by the mWGS approach (Table 1; Supplementary Figure S2).
BLASTn searches were used to match OTUs derived from the TEFAP approach to unprocessed mWGS reads. 57.14% of C. concentrica KS domain OTUs and 29.41% of R. odorabile KS domain OTUs matched to the mWGS sequences. Only 0.62% of C. coralliophila and 2.15% of Scopalina sp. C domain OTUs were matched to mWGS sequences. No overlap was observed for C domain sequences from C. concentrica or KS domain sequences from Scopalina sp., C. coralliophila and Stylissa sp. In order to scrutinise this lack of overlap, the taxonomic identity of reads from both the TEFAP and mWGS data sets were compared. Distinct differences were observed between the two methods in regard to the confidence at which sequences were assigned to more resolved taxonomic groups (see Supplementary Figure S3). Furthermore, the TEFAP approach enriched for sequences with GC contents ranging between 45 and 50%, and 60 and 65%, while the mWGS data showed a dominance of sequences with GC contents ranging between 25 and 35% for C domains and 65 and 75% for KS domains (see Supplementary Figure S4).
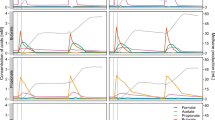
Reads derived from the TEFAP methods were used to calculate diversity and richness indices for each sponge where at least 324 reads were obtained. As a result of low sequence coverage, only three sponges could be analysed for their KS domains and two for their C domain sequences. Overall, the C domains presented greater observed richness (ACE), whereas the KS domains had higher observed diversity (invSimpson) (Table 2). KS and C domain richness and diversity were highest within the sponge Scopalina sp. Despite being dominated by a single functional group of KS domains (Figure 1), R. odorabile contained a more diverse population of KS domains compared with C. concentrica.
The TEFAP approach and mWGS revealed different phylogenetic compositions of C and KS domains with the marine sponges (Figure 1 and Figure 2). The two sponges C. concentrica and Scopalina sp. were shown, by both methods, to contain C domains responsible for the condensation of two L-amino subunits, annotated as symbiont LCL Clade 1 and Symbiont LCL Clade 2, that contained only sequences obtained from this study (Figure 2). The specific taxonomic assignment of sequences to these clades varied between the two methods. Both methods were also in agreement as to the presence of a clade of proteobacteria-like LCL domain sequences in the sponge C. concentrica and the presence of cyanobacteria-like LCL domain sequences in the sponge Scopalina sp. The TEFAP method alone revealed the presence of Proteobacteria-like LCL sequences in Scopalina, and cyanobacteria-like LCL in C. concentrica and C. coralliophila.
Among KS domains a similar pattern was observed (Figure 1). Both methods were in agreement as to the dominance of hybrid NRPS/PKS sequences in C. concentrica, trans-AT sequences in Scopalina sp. and sponge ubiquitous PKS (sup) sequences in R. odorabile. In each of these three instances, additional phylogenetic groups were observed when the sponge was surveyed by the TEFAP approach. The mWGS approach was able to identify hybrid NRPS/PKS sequences within the sponge C. coralliophila, though these sequences were not detected using the TEFAP approach.
Discussion
Amplicon pyrosequencing enables unprecedented discovery of natural product biosynthesis
In this study, a TEFAP approach targeting natural product biosynthetic genes was applied to six marine sponge species. This study reports the use of a TEFAP approach for simultaneously evaluating the diversity of multiple domains involved in natural product biosynthesis from a number of environments. Both NRPS and PKS biosynthetic pathways were detected within the microbiomes of Scopalina sp., C. coralliophila and C. concentrica, whereas only PKS biosynthetic pathways were identified within the microbiomes of Stylissa sp. and R. odorabile. The TEFAP approach resulted in the identification of ∼100 novel KS domain and 400 novel C domain sequences. While there was an apparent lack of coverage for these domains in some of the sponge samples, overall the diversity among the Australian sponge species as revealed by the TEFAP approach exceeded that revealed by mWGS. In addition the diversity of KS and C domains within the microbiomes of individual sponges, particular Scopalina sp., was greater than that of any other marine sponge (Fieseler et al., 2007; Kennedy et al., 2008; Pimentel-Elardo et al., 2012). This is despite a relaxed clustering approach, whereby a 0.10 distance threshold was adopted (Varaljay et al., 2010; Howard et al., 2011) for the identification of novel sequences, in contrast to the 97% nucleotide similarity cutoff reported in comparable studies (Fieseler et al., 2007; Pimentel-Elardo et al., 2012). The relaxed cutoff would also account for any inflation of the discovery rate due to random sequencing errors. Within individual samples, the TEFAP and mWGS methods applied were in some disagreement in regards to the average GC content (see Supplementary Figure S4), as well as the functional (Figures 1 and 2) and taxonomic (see Supplementary Figure S3) composition. This would indicate a potential technical biases of amplicon-dependent approaches leading to the artificial enrichment of certain sequences as observed by others (Piel, 2002; Piel et al., 2004a; Fisch et al., 2009; Pimentel-Elardo et al., 2012).
The occurrence of technical biases associated with the TEFAP method implies that the reported relative composition of KS and C domain sequences within each sample is skewed. The most notable implication is the failure to detect KS and C domains corresponding to the high GC Gram-positive actinobacteria and firmicutes. This is directly reflected within GC plots (see Supplementary Figure S4), with the TEFAP method selecting for two GC ranges, likely corresponding to cyanobacterial and proteobacterial groups (Figures 1 and 2). Ultimately, this suggests that the diversity of both KS and C domain sequences within a sponge is possibly higher than that observed.
An additional consideration is that of modular redundancy, whereby multiple C and KS domains cooperate within a single biosynthetic pathway. Efforts to avoid excessive redundancy were made by utilising the C domain in favour of the commonly applied A domain, an approach, which has already been adopted by others (Ziemert et al., 2012). In addition, we adopted a low distance threshold to account for gene duplications that may give rise to multimodular systems. However, the assembly-line logic of these biosynthetic pathways stipulates that multiple genetically distinct C and KS modules will be present in a single pathway leading to overestimations of diversity. Although one could assume that typical pathways feature 3–7 condensing domains per biosynthethic pathway, targeted recovery of large metagenomic fragments is ultimately required to determine the extent of this redundancy (Piel et al., 2004a; Fisch et al., 2009). Regardless of these considerations, ensuring that multiple environments are sampled using a standardised approach, the TEFAP method allows for comparative analysis of composition and diversity between multiple environments.
A novel TEFAP approach highlights the high diversity of PKS genes in the microbiomes of Australian marine sponges
This study represents the first analysis of PKS diversity from Australian marine sponges. A previous study, utilising vector-based approaches, reported the identification of 150 unique KS OTUs (97% nucleotide identity) that were derived by undertaking Sanger sequencing of nearly 500 amplicons from the three marine sponges Theonella swinhoei, Cacospongia mycofijiensis and Aplysina aerophoba (Fieseler et al., 2007). In contrast, TEFAP enabled the discovery of nearly 100 KS domain sequences from the five Australian marine sponges, albeit at a lower distance threshold of 0.10. At a comparable distance threshold of 0.03, the TEFAP method reported the discovery of 296 OTUs (data not shown), far exceeding previous vector-based studies (Fieseler et al., 2007) but comparable with other amplicon sequencing approaches (Trindade-Silva et al., 2012).
Schirmer et al. (2005) and Fieseler et al. (2007) both reported the dominance of sup type KS domains within amplicon clone libraries from Discoderma dissoluta, Theonella swinhoei, Cacospongia mycofijiensis and Aplysina aerophoba, as well as the detection sup type KS in Verongula gigantea, Aiolochroia crassa, Xestospongia muta and Siphonodictyon corralliphagum. In this study, sup KS domains were detected in abundance within the sponge R. odorabile, along with a small number of cis-KS domains phylogenetically affiliated with actinobacteria. This co-occurrence of these two domain types was also observed from the sponge Cacospongia mycofijiensis (Fieseler et al., 2007; Hochmuth et al., 2010), which belongs to the same order as R. odorabile. The occurrence of these sup type KS domains in R. odorabile coincided with the detection of poribacteria (Fan et al., 2012), which are known to harbour the sup biosynthetic gene cluster (Siegl and Hentschel, 2010; Siegl et al., 2011). The calculated richness index (Table 2), indicated a moderate number of sup KS domains within the microbiome of this sponge. That this moderate richness corresponded with a low invSimpson value, indicating that a few sup domains were dominant, reflects the shallow phylogenetic branching nature of this group (Fieseler et al., 2007; Hochmuth and Piel, 2009). In contrast to R. odorabile, poribacteria were not previously detected in the sponges C. coralliophila and Stylissa sp. (Fan et al., 2012) using the mWGS technique. However, the TEFAP approach detected sup type KS domains, which are thought to be limited to poribacterial species (Hochmuth and Piel, 2009; Siegl and Hentschel, 2010; Siegl et al., 2011). The detection of sup domains at very low abundance has also been reported within the marine sponge Arenosclera brasiliensis (Trindade-Silva et al., 2012). This discrepancy may be explained by poribacteria constituting only a small proportion of the sponge microbiome in these samples, reflective of levels observed in seawater (Taylor et al., 2012). As such the level of sequencing achieved in the mWGS approach (Fan et al., 2012), which targets the most abundant taxa, may not have been sufficient to retrieve rare sequences. However, it is possible that the sup type KS domain is not exclusive to poribacteria, but is present in other lineages.
While Fieseler et al. (2007) reported that sup type KS domains dominated the majority of sponges screened, the presence of trans-acyltransferase (trans-AT) type KS domains are more relevant for the discovery of novel bioactive small molecules. Previously identified pathways containing trans-AT domains are responsible for the biosynthesis of complex polyketides with bioactivies relevant to the pharmaceutical industry (Piel, 2002; Piel et al., 2004b; Fisch et al., 2009). The trans-AT KS domains, detected in this study within the sponges C. coralliophila, Scopalina sp. and Stylissa sp. (Figure 1), are distinct from typical Type I or cis-AT KS in that one of the core catalytic domains is absent. In these systems, the AT domain, responsible for activation of the carboxylic acid precursor, is found as a mono-functional enzyme proximal to the biosynthetic gene cluster. To date, a number of trans-AT biosynthetic pathways have been reported primarily from organisms in symbiotic associations (Piel, 2002; Piel et al., 2004b; Fisch et al., 2009). Scopalina sp. contained 41 KS domains, the majority of which were trans-AT type. The trans-AT domains from Scopalina sp., C. coralliophila and Stylissa, did not exhibit >90% amino-acid identity to any known biosynthetic pathways, indicating the presence of novel biosynthetic pathways within these sponges. Surprisingly, a survey of the literature revealed a lack of bioactive compounds isolated from Scopalina spp., which in the context of this study may represent a large untapped resource for natural product discovery.
Metagenomic mining of condensation domains reveals an unprecedented diversity of NRP biosynthesis
Despite the large chemical diversity of cyclic peptides isolated from sponges and other marine invertebrates (Fusetani and Matsunaga, 1993; De Rosa et al., 2003; Thomas et al., 2010), C domains were only detected within the microbial communities of half the surveyed sponges. A high richness was observed among C domains within the sponges Scopalina sp. and C. coralliophila, particularly in contrast to that observed among KS domains (Table 2). The higher invSimpson values, among the C domain sequences suggested a more even distribution with far fewer rare sequences. This is likely to reflect either the presence of a single super-producing organism, or indicate that many of these C domains are present in a small number of clusters. The phylogenetic inference of the C domains is enabled by the presence of two binding pockets adjacent to the catalytic centre (Stachelhaus et al., 1998). The C-terminus binding pocket, considered the acceptor site, exhibits specific selectivity for the activated substrate, which is conferred within the amino-acid sequence. This allows for the discrimination between activated L-amino, D-amino or N-acyl substrates (Roongsawang et al., 2005; Rausch et al., 2007). Within this study, both the TEFAP and mWGS approaches were in agreement as to the presence of only LCL domains within Scopalina sp., C. concentrica and C. coralliophila. These domain types are defined by the presence of an L-amino acid in both the acceptor and donor binding pocket (Roongsawang et al., 2005; Rausch et al., 2007). Further phylogenetic analysis revealed both methods were in agreement that these LCL domains were limited to Gram-negative organisms, particularly members of the γ-proteobacteria, δ-proteobacteria and cyanobacteria (Figure 2).
Despite not detecting cyanobacteria within Scopalina sp. and C. concentrica (Fan et al., 2012), the TEFAP approach amplified reads that form a phylogenetic clade with C domains from the marine cyanobacteria Moorea producens (Jones et al., 2011) and Acaryochloris marina (Swingley et al., 2008). Within Scopalina sp. and C. concentrica, a number of sequences also formed a phylogenetic clade with C domains from proteobacteria (Figure 2). Proteobacteria LCL Clade 3 contained exclusively reference sequences from members of the γ-proteobacteria, a group that was not identified from mWGS of Scopalina sp. and C. concentrica. For Scopalina sp., this may indicate the presence of γ-proteobacteria in the ‘rare biosphere’ (Sogin et al., 2006). However, in the instance of C. concentrica, the mWGS method also identified sequences that grouped within proteobacteria LCL Clade 3, suggesting that in this case the organism harbouring this pathway is most likely abundant.
A large number of C domain sequences from the two marine sponges, C. concentrica and Scopalina sp., as surveyed by the TEFAP method, formed a phylogenetic clade independent of any reference sequences (Figure 2). A second phylogenetic clade containing exclusively mWGS derived sequences was also observed. These clades were annotated as Symbiont LCL Clade 1 and 2, respectively. The presence of these two phylogenetic clades, comprised entirely of C domains from sponge symbionts, is particularly remarkable considering that previous analyses of KS from marine sponges have revealed distinct taxonomic groups (Piel et al., 2004b; Trindade-Silva et al., 2012), leading to the definition of, in one instance, the sup type KS group (Fieseler et al., 2007).
Conclusions
Bioactive natural products of NRPS and PKS origin are highly valued by the pharmaceutical industry as lead compounds against existing and emerging diseases. Researchers within the natural product discipline are under increasing pressures to find new resources for these compounds. During the last 10 years, significant focus has been placed on screening environments in an effort to identify natural products produced by organisms intractable to traditional methods. The approaches applied to date have been successful, however, these technologies have always been applied to environments where there is sufficient prior knowledge regarding natural product diversity. In this study, we have highlighted the untapped potential of Australian marine sponges, in particular that of the genus Scopalina sp., for the discovery of natural products. Furthermore, we have done so using a novel TEFAP approach that allows for the screening of multiple environments simultaneously. Although, it is clear from comparisons with mWGS data that the TEFAP approach has some bias regarding the relative composition of particular sequences. The TEFAP method is superior to other amplicon-dependent methods, in that the generation of substantially larger numbers of sequences accounts for this intrinsic bias, while providing sufficient information for the targeted recovery of large metagenomic regions from cloned genomic libraries. In this study, we have highlighted how such an approach can reveal, and provide access to, the biosynthetic potential of previously uncharacterised environments. Following from this, it is envisaged that this approach can be applied to multiple different ecosytems in order to present a picture of how these environments select for the presence of organisms producing natural products.
References
Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W et al (1997). Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25: 3389–3402.
Banik JJ, Brady SF . (2010). Recent application of metagenomic approaches toward the discovery of antimicrobials and other bioactive small molecules. Curr Opin Microbiol 13: 603–609.
Benson DA, Boguski MS, Lipman DJ, Ostell J . (1997). GenBank. Nucleic Acids Res 25: 1–6.
Blunt JW, Copp BR, Munro MHG, Northcote PT, Prinsep MR . (2006). Marine natural products. Nat Prod Rep 23: 26–78.
Boot CM, Tenney K, Valeriote FA, Crews P . (2006). Highly N-methylated linear peptides produced by an atypical sponge-derived Acremonium sp. J Nat Prod 69: 83–92.
Brady SF, Simmons L, Kim JH, Schmidt EW . (2010). Metagenomic approaches to natural products from free-living and symbiotic organisms. Nat Prod Rep 26: 1488–1503.
Chiang Y-M, Chang S-L, Oakley BR, Wang CC . (2010). Recent advances in awakening silent biosynthetic gene clusters and linking orphan clusters to natural products in microorganisms. Curr opin Chem Biol 15: 137–143.
Craig JW, Chang F-Y, Kim JH, Obiajulu SC, Brady SF . (2010). Expanding small-molecule functional metagenomics through parallel screening of broad-host-range cosmid environmental DNA libraries in diverse Proteobacteria. Appl Environ Microbiol 76: 1633–1641.
De Rosa S, Mitova M, Tommonaro G . (2003). Marine bacteria associated with sponge as source of cyclic peptides. Biomol Eng 20: 311–316.
Donachie SP, Foster JS, Brown MV . (2007). Culture clash: challenging the dogma of microbial diversity. ISME J 1: 97–99.
Edgar RC . (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32: 1792–1797.
Fan L, Reynolds D, Liu M, Stark M, Kjelleberg S, Webster N et al (2012). Functional equivalence and evolutionary convergence in complex communities of microbial symbionts. Proc Natl Acad Sci USA 109: E1878–E1887.
Ferrari BC, Winsley T, Gillings M, Binnerup S . (2008). Cultivating previously uncultured soil bacteria using a soil substrate membrane system. Nat Protoc 3: 1261–1269.
Fieseler L, Hentschel U, Grozdanov L, Schirmer A, Wen G, Platzer M et al (2007). Widespread occurrence and genomic context of unusually small polyketide synthase genes in microbial consortia associated with marine sponges. Appl Environ Microbiol 73: 2144–2155.
Finn RD, Clements J, Eddy SR . (2011). HMMER web server: interactive sequence similarity searching. Nucleic Acids Res 39: W29–W37.
Finn RD, Mistry J, Tate J, Coggill P, Heger A, Pollington JE et al (2010). The Pfam protein families database. Nucleic Acids Res 38: D211–D222.
Fisch KM, Gurgui C, Heycke N, van der Sar SA, Anderson SA, Webb VL et al (2009). Polyketide assembly lines of uncultivated sponge symbionts from structure-based gene targeting. Nat Chem Biol 5: 494–501.
Fu J, Wenzel SC, Perlova O, Wang J, Gross F, Tang Z et al (2008). Efficient transfer of two large secondary metabolite pathway gene clusters into heterologous hosts by transposition. Nucleic Acids Res 36: e113.
Fusetani N, Matsunaga S . (1993). Bioactive sponge peptides. Chem Rev 93: 1793–1806.
Guindon S, Lethiec F, Duroux P, Gascuel O . (2005). PHYML Online—a web server for fast maximum likelihood-based phylogenetic inference. Nucleic Acids Res 33: W557–W559.
Hochmuth T, Niederkruger H, Gernert C, Siegl A, Taudien S, Platzer M et al (2010). Linking chemical and microbial diversity in marine sponges: possible role for poribacteria as producers of methyl-branched fatty acids. Chembiochem 11: 2572–2578.
Hochmuth T, Piel J . (2009). Polyketide synthases of bacterial symbionts in sponges—evolution-based applications in natural products research. Phytochemistry (Oxf) 70: 1841–1849.
Howard EC, Sun S, Reisch CR, del Valle DA, Bürgmann H, Kiene RP et al (2011). Changes in dimethylsulfoniopropionate demethylase gene assemblages in response to an induced phytoplankton bloom. Appl Environ Microbiol 77: 524–531.
Huson DH, Mitra S, Ruscheweyh HJ, Weber N, Schuster SC . (2011). Integrative analysis of environmental sequences using MEGAN4. Genome Res 21: 1552–1560.
Jones AC, Monroe EA, Podell S, Hess WR, Klages S, Esquenazi E et al (2011). Genomic insights into the physiology and ecology of the marine filamentous cyanobacterium Lyngbya majuscula. Proc Natl Acad Sci USA 108: 8815–8820.
Keller M, Zengler K . (2004). Tapping into microbial diversity. Nat Rev Microbiol 2: 141–150.
Kennedy J, Codling CE, Jones BV, Dobson ADW, Marchesi JR . (2008). Diversity of microbes associated with the marine sponge, Haliclona simulans, isolated from Irish waters and identification of polyketide synthase genes from the sponge metagenome. Environ Microbiol 10: 1888–1902.
Moffitt MC, Neilan BA . (2003). Evolutionary affiliations within the superfamily of ketosynthases reflect complex pathway associations. J Mol Evol 56: 446–457.
Newman DJ, Cragg GM . (2012). Natural products as sources of new drugs over the 30 years from 1981 to 2010. J Nat Prod 75: 311–335.
Piel J . (2002). A polyketide synthase-peptide synthetase gene cluster from an uncultured bacterial symbiont of Paederus beetles. Proc Natl Acad Sci USA 99: 14002–14007.
Piel J, Hui D, Fusetani N, Matsunaga S . (2004a). Targeting modular polyketide synthases with iteratively acting acyltransferases from metagenomes of uncultured bacterial consortia. Environ Microbiol 6: 921–927.
Piel J, Hui D, Wen G, Butzke D, Platzer M, Fusetani N et al (2004b). Antitumor polyketide biosynthesis by an uncultivated bacterial symbiont of the marine sponge Theonella swinhoei. Proc Natl Acad Sci USA 101: 16222–16227.
Pimentel-Elardo SM, Grozdanov L, Proksch S, Hentschel U . (2012). Diversity of nonribosomal peptide synthetase genes in the microbial metagenomes of marine sponges. Mar Drugs 10: 1192–1202.
Rausch C, Hoof I, Weber T, Wohlleben W, Huson D . (2007). Phylogenetic analysis of condensation domains in NRPS sheds light on their functional evolution. BMC Evol Biol 7: 78.
Rice P, Longden I, Bleasby A . (2000). EMBOSS: the European molecular biology open software suite. Trends Genet 16: 276–277.
Roongsawang N, Lim SP, Washio K, Takano K, Kanaya S, Morikawa M . (2005). Phylogenetic analysis of condensation domains in the nonribosomal peptide synthetases. FEMS Microbiol Lett 252: 143–151.
Schirmer A, Gadkari R, Reeves CD, Ibrahim F, Delong EF, Hutchinson CR . (2005). Metagenomic Analysis Reveals Diverse Polyketide Synthase Gene Clusters in Microorganisms Associated with the Marine Sponge Discodermia dissoluta. Society 71: 4840–4849.
Schloss PD, Westcott SL . (2011). Assessing and improving methods used in operational taxonomic unit-based approaches for 16S rRNA gene sequence analysis. Appl Environ Microbiol 77: 3219–3226.
Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB et al (2009). Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 75: 7537–7541.
Schmitt I, Kautz S, Lumbsch HT . (2008). 6-MSAS-like polyketide synthase genes occur in lichenized ascomycetes. Mycol Res 112: 289–296.
Siegl A, Hentschel U . (2010). PKS and NRPS gene clusters from microbial symbiont cells of marine sponges by whole genome amplification. Environ Microbiol Rep 2: 507–513.
Siegl A, Kamke J, Hochmuth T, Piel J, Richter M, Liang C et al (2011). Single-cell genomics reveals the lifestyle of Poribacteria, a candidate phylum symbiotically associated with marine sponges. ISME J 5: 61–70.
Sogin ML, Morrison HG, Huber JA, Welch DM, Huse SM, Neal PR et al (2006). Microbial diversity in the deep sea and the underexplored ‘rare biosphere’. Proc Natl Acad Sci USA 103: 12115–12120.
Stachelhaus T, Mootz HD, Bergendahl V, Marahiel MA . (1998). Peptide bond formation in nonribosomal peptide biosynthesis catalytic role of the condensation domain. J Biol Chem 273: 22773–22781.
Sun Y, Wolcott RD, Dowd SE . (2011). Tag-encoded FLX amplicon pyrosequencing for the elucidation of microbial and functional gene diversity in any environment. Methods Mol Biol 733: 129–141.
Swingley WD, Chen M, Cheung PC, Conrad AL, Dejesa LC, Hao J et al (2008). Niche adaptation and genome expansion in the chlorophyll d-producing cyanobacterium Acaryochloris marina. Proc Natl Acad Sci USA 105: 2005–2010.
Taylor MW, Tsai P, Simister RL, Deines P, Botte E, Ericson G et al (2012). ‘Sponge-specific’bacteria are widespread (but rare) in diverse marine environments. ISME J 7: 438–443.
Thomas TRA, Kavlekar DP, LokaBharathi PA . (2010). Marine drugs from sponge-microbe association—a review. Mar Drugs 8: 1417–1468.
Trindade-Silva AE, Rua CP, Andrade BG, Vicente AC, Silva GG, Berlinck RG et al (2012). Polyketide synthase gene diversity within the endemic sponge Arenosclera brasiliensis microbiome. Appl Environ Microbiol 79: 1598–1605.
Varaljay VA, Howard EC, Sun S, Moran MA . (2010). Deep sequencing of a dimethylsulfoniopropionate-degrading gene (dmdA) by using PCR primer pairs designed on the basis of marine metagenomic data. Appl Environ Microbiol 76: 609–617.
Walsh CT, Fischbach MA . (2010). Natural products version 2.0: connecting genes to molecules. J Am Chem Soc 132: 2469–2493.
Ziemert N, Podell S, Penn K, Badger JH, Allen E, Jensen PR . (2012). The Natural Product Domain Seeker NaPDoS: A Phylogeny Based Bioinformatic Tool to Classify Secondary Metabolite Gene Diversity. PLoS ONE 7: e34064.
Acknowledgements
We would like to acknowledge the financial support from the Australian Research Council and the Gordon and Betty Moore Foundation.
Author Contributions
JNW, MVB and BAN designed research; JNWand LF performed research; JNW, MVB, LF, TT and BAN analysed data; and JNW, MVB, TT and BAN wrote the manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on The ISME Journal website
Rights and permissions
About this article
Cite this article
Woodhouse, J., Fan, L., Brown, M. et al. Deep sequencing of non-ribosomal peptide synthetases and polyketide synthases from the microbiomes of Australian marine sponges. ISME J 7, 1842–1851 (2013). https://doi.org/10.1038/ismej.2013.65
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ismej.2013.65
Keywords
This article is cited by
-
Next Generation High Throughput Sequencing to Assess Microbial Communities: An Application Based on Water Quality
Bulletin of Environmental Contamination and Toxicology (2021)
-
Localized production of defence chemicals by intracellular symbionts of Haliclona sponges
Nature Microbiology (2019)
-
A computationally simplistic poly-phasic approach to explore microbial communities from the Yucatan aquifer as a potential sources of novel natural products
Journal of Microbiology (2016)
-
Culture-independent discovery of natural products from soil metagenomes
Journal of Industrial Microbiology and Biotechnology (2016)
-
The integration of sequencing and bioinformatics in metagenomics
Reviews in Environmental Science and Bio/Technology (2015)