Abstract
Coevolution with phages drive the evolution of high bacterial mutation rates in vitro, but the relevance of this finding to natural populations is unclear. Here, we investigated how coevolution affects mutation rate evolution in soil, in the presence and absence of the rest of the natural microbial community. Although mutation rate on average increased threefold, neither coevolving phages nor the rest of natural community significantly affected mutation rates. Our results suggest that features of the soil over and above directly interacting organisms constrain the evolution of strong mutators, helping to explain their relatively low frequency compared with some laboratory and clinical settings.
Similar content being viewed by others
Main
Mutator bacteria readily evolve in experimental populations (Sniegowski et al., 1997; Giraud et al., 2001; Pal et al., 2007), and are found at relatively high frequencies in both natural populations (LeClerc et al., 1996; Matic et al., 1997) and in clinical opportunistic pathogen populations (Oliver et al., 2000). Although mutators may increase adaptability to novel environmental conditions (Sniegowski et al., 1997; Giraud et al., 2001; Pal et al., 2007), they are also prone to the accumulation of deleterious mutations (McDonald et al., 2012). The long-term maintenance of high mutation rates requires rapidly changing selection pressures (Tenaillon et al., 1999; Denamur and Matic, 2006; Desai and Fisher, 2011), in addition to the possible slow transition rate by point mutation from mutators to non-mutators (Denamur et al., 2000). One of the most likely causes of rapidly changing selection pressures is coevolution with lytic bacteriophages (Pal et al., 2007). Lytic bacteriophages are ubiquitous, and require bacterial cell lysis following infection and replication to transmit to new hosts, hence there is very strong reciprocal selection for bacterial resistance and phage infectivity. This interaction can lead to ongoing antagonistic coevolution between bacteria and phages (Bohannan and Lenski, 2000; Buckling and Rainey, 2002); hence, creating conditions where mutator alleles may increase in frequency by hitch-hiking with the beneficial resistance mutations they generate.
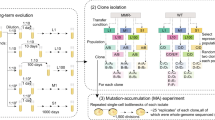
Recent in vitro work has shown that coevolution between the bacterium Pseudomonas fluorescens SBW25 (Rainey and Bailey, 1996) and a lytic dsDNA phage (Buckling and Rainey, 2002) rapidly results in the evolution of elevated mutations rates, and competition between wild-type and isogenic mutator bacteria confirm the selective advantage in the presence but not the absence of coevolving phages (Pal et al., 2007). However, the relevance of this finding to natural populations is unclear. Although this bacteria and phage readily coevolve in their natural environment, soil (Gómez and Buckling, 2011), phage-imposed selection for resistance is lower in soil than in vitro despite long-term persistence of the phage population (Buckling and Rainey, 2002; Gómez and Buckling, 2011). Moreover, selection for mutation rates imposed by a single phage may be unimportant when experiencing the myriad selection pressures resulting from the community as a whole. Here, we determine the simultaneous roles of a tightly coevolving phage and interactions with the rest of the natural microbial and viral community (NMC) in driving the evolution of mutation rates of P. fluorescens in soil. We inoculated 32 sterile soil microcosms; 10 × 10 cm2 petri dishes containing 100 g of twice autoclaved (unsieved) compost (John Innes no. 2), with P. fluorescens SBW25, half of which were then inoculated with a soil wash of the NMC, and half with phage SBW25, in a fully factorial design, as described previously in Gómez and Buckling (2011).
After 48 days evolution in soil, we determined the mutation rates of P. fluorescens SBW25 using simple fluctuation tests (Luria and Delbruck, 1943). Twenty bacteria colonies of P. fluorescens SBW25 were taken randomly from each of the 32 populations, and we grew all the colonies from each population together King’s media B (KB) overnight. Four new cultures of each of the ancestral bacteria and the 32 evolved bacterial cultures were established with 100 cells from a previous culture, and grown for 24 h at 28 °C in a shaken (200 r.p.m.) incubator, before plating onto both normal KB agar and KB agar supplemented with rifampicin (100 μg ml−1). Bacterial mutation rates were estimated by fluctuation tests for antibiotic resistance (Luria and Delbruck, 1943), having previously established that no colonies from the evolved populations in the soil microcosms were resistant to rifampicin. Mutation rates were calculated and corrected for sampling error using the Ma–Sandri–Sarkar maximum likelihood estimator (MSS-MLE) method (Rosche and Foster, 2000), implemented in the FALCOR web package (Hall et al., 2009).
P. fluorescens showed an average threefold increase in mutation rates (Figure 1; one sample t-test for all populations against ancestral: t31=6.04, P<0.01), perhaps because the specific conditions represents a slightly novel and stressful environment for P. fluorescens. However, populations evolved in the presence of phages did not evolve higher mutation rates than populations evolved in the absence (F1,28=4.1; P=0.06); if anything, the presence of phage resulted in lower mutation rates. The absence of a clear difference in mutation rates between treatments cannot be explained by the absence of phage-imposed selection in the phage treatments: phage persisted throughout the experiment (Gómez and Buckling, 2011), and some bacteria exposed to phages evolved phage resistance to ancestral and sympatric phages, whereas resistance was undetected in the phage-free treatments (Supplementary Figure S1). Moreover, the presence of the NMC or the interaction between phages and the NMC had no impact on mean mutation rates (Figure 1; P>0.2 in both cases).
Mean (±s.e.m.) mutation rates of the ancestral P. fluorescens SBW25 bacteria (black bar) and evolved bacteria populations isolated after 48 days evolving with (+P; grey bar) and without phages (−P; white bar) and in the absence (−NMC) and presence (+NMC) of the natural microbial community. Mutation rate was determined by fluctuation test to rifampicin resistance and calculated by The Ma–Sandri–Sarkar maximum likelihood estimator (MSS-MLE) method. For evolved bacteria, each bar represents the average of eight independent populations from four replicates per population.
The likelihood of mutators successfully invading populations is increased by population size (which increases the chance of mutators generating beneficial mutations and thus hitch-hiking to high frequencies), time and the strength of selection (Tenaillon et al., 1999). We suggest that population size cannot explain the absence of phage-imposed increases in mutation rate because P. fluorescens population densities were extremely large (∼107 g−1 soil; Gómez and Buckling, 2011), and only 10–100-fold less than that observed in vitro (Buckling and Hodgson, 2007). Although we do not know the generation time of bacteria in soil, it is inevitably longer than in a media, hence the absence of a large change in mutation rate resulting from phage-imposed selection may have resulted from a time constraint of the experiment: strong mutators may not have had a chance to reach high frequencies. To investigate this possibility, we competed a lacZ-marked of P. fluorescens SBW25 (wild-type) against a mutL-knockout mutant of P. fluorescens SBW25 (a 100-fold mutator) (Pal et al., 2007) in the presence and the absence of phages for 10 days in soil—a period long enough for resistance evolution to occur and hence for any relative advantage of mutators in the absence versus the presence of phages to be uncovered. Mutators suffered a general competitive cost in soil (t11=3.69, P<0.01 in the absence of any genetic marker; Pal et al., 2007), possibly due to pleiotropic effects of the knockout, and that was not influenced by phages (Figure 2; F1,10=3.453; P=0.09). We can therefore conclude that phage simply do not impose sufficiently strong selection on bacteria in soil for mutator alleles to hitch-hike to detectable frequencies.
Relative fitness of the P. fluorescens SBW25 mutator (mutL) bacteria to the wild-type bacteria in the presence (+P; grey bar) and the absence (−P; white bar) of phages after 10 days of competition in soil. Bars represent mean (±s.e.m.) relative fitness (n=6). Note that if relative fitness=1, mutator and wild-type bacteria are equally fit.
Why do phages select for large increases in vitro but not in vivo, having ruled out the impact of the rest of natural microbial community? Numerical simulations (Pal et al., 2007) suggest a major factor affecting the evolution of mutators when coevolving with phages is the cost associated with phage resistance: when costs are high, there is weaker selection both for resistance and the mutator alleles that generate resistance mutations. Crucially, costs of resistance are much greater in soil than in vitro (Gómez and Buckling, 2011). Moreover, the coevolutionary dynamics between bacteria and virus are qualitatively different in soil compared with in vitro: coevolution in soil is characterised by fluctuations in resistance and infectivity through space and time (Gómez and Buckling, 2011), whereas coevolution in vitro follows and arms race dynamic (Gandon et al., 2008), resulting in bacteria and phages evolving increasingly wide resistance and infectivity ranges through time, respectively (Buckling and Rainey, 2002).
In summary, direct evolutionary interactions with viruses and the other members of soil microbial communities appear to have relatively minor roles in driving the evolution of bacterial mutators in soil. Other features of the soil environment appear to select for relatively low mutation rates, as confirmed by the fitness cost of the 100-fold mutator in the competition experiments. We cautiously generalise the results from this experimental system on the basis that spatial patterns of phage adaption to bacteria in natural soil communities are consistent with rapid bacteria–virus coevolution being the norm (Vos et al., 2009). These results may therefore help to explain why bacteria with high mutation rates (10–1000-fold higher than the wild-type) are found at relatively low frequencies (<2%) in soil, but can be found at much higher frequencies in the lab and in clinical infections.
References
Bohannan B, Lenski R . (2000). The relative importance of competition and predation varies with productivity in a model community. Am Nat 156: 329–340.
Buckling A, Hodgson DJ . (2007). Short-term rates of parasite evolution predict the evolution of host diversity. J Evol Biol 20: 1682–1688.
Buckling A, Rainey PB . (2002). Antagonistic coevolution between a bacterium and a bacteriophage. P Roy Soc B Biol Sci 269: 931–936.
Denamur E, Lecointre G, Darlu P, Tenaillon O, Acquaviva C, Sayada C et al (2000). Evolutionary implications of the frequent horizontal transfer of mismatch repair genes. Cell 103: 711–721.
Denamur E, Matic I . (2006). Evolution of mutation rates in bacteria. Mol Microbiol 60: 820–827.
Desai MM, Fisher DS . (2011). The balance between mutators and nonmutators in asexual populations. Genetics 188: 997–1014.
Gandon S, Buckling A, Decaestecker E, Day T . (2008). Host-parasite coevolution and patterns of adaptation across time and space. J Evol Biol 21: 1861–1866.
Giraud A, Matic I, Tenaillon O, Clara A, Radman M, Fons M et al (2001). Costs and benefits of high mutation rates: adaptive evolution of bacteria in the mouse gut. Science 291: 2606–2608.
Gómez P, Buckling A . (2011). Bacteria-phage antagonistic coevolution in soil. Science 332: 106–109.
Hall BM, Ma C-X, Liang P, Singh KK . (2009). Fluctuation analysis calculator: a web tool for the determination of mutation rate using Luria-Delbruck fluctuation analysis. Bioinformatics 25: 1564–1565.
LeClerc JE, Li B, Payne WL, Cebula TA . (1996). High mutation frequencies among Escherichia coli and Salmonella pathogens. Science 274: 1208–1211.
Luria SE, Delbruck M . (1943). Mutations of bacteria from virus sensitivity to virus resistance. Genetics 28: 491–511.
Matic IM, Radman F, Taddei B, Picard C, Doit E, Bingen E et al (1997). Highly variable mutation rates in commensal and pathogenic Escherichia coli. Science 277: 1833–1834.
McDonald MJ, Hsieh YY, Yu YH, Chang SL, Leu JY . (2012). The evolution of low mutation rates in experimental mutator populations of Saccharomyces cerevisiae. Curr Biol 22: 1235–1240.
Oliver A, Canton R, Campo P, Baquero F, Blazquez J . (2000). High frequency of hypermutable Pseudomonas aeruginosa in cystic fibrosis lung infection. Science 288: 1251–1253.
Pal C, Macia MD, Oliver A, Schachar I, Buckling A . (2007). Coevolution with viruses drives the evolution of bacterial mutation rates. Nature 450: 1079–1081.
Rainey PB, Bailey MJ . (1996). Physical and genetic map of the Pseudomonas fluorescens SBW25 chromosome. Mol Microbiol 19: 521–533.
Rosche WA, Foster PL . (2000). Determining mutation rates in bacterial populations. Methods 20: 4–17.
Sniegowski PD, Gerrish PJ, Lenski RE . (1997). Evolution of high mutation rates in experimental populations of E-coli. Nature 387: 703–705.
Tenaillon O, Toupance B, Le Nagard H, Taddei F, Godelle B . (1999). Mutators, population size, adaptive landscape and the adaptation of asexual populations of bacteria. Genetics 152: 485–493.
Vos M, Birkett PJ, Birch E, Griffiths RI, Buckling A . (2009). Local adaptation of bacteriophages to their bacterial hosts in soil. Science 325: 833.
Acknowledgements
We thank two anonymous reviews for comments on the manuscript. The work was supported by European Research Council. PG was supported by a Marie Curie Intra-European Fellowship (PIEF-GA-2010-272945) within the European Union Seventh Framework Programme.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on The ISME Journal website
Supplementary information
Rights and permissions
About this article
Cite this article
Gómez, P., Buckling, A. Coevolution with phages does not influence the evolution of bacterial mutation rates in soil. ISME J 7, 2242–2244 (2013). https://doi.org/10.1038/ismej.2013.105
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ismej.2013.105
Keywords
This article is cited by
-
Interactions between bacterial and phage communities in natural environments
Nature Reviews Microbiology (2022)
-
Community context matters for bacteria-phage ecology and evolution
The ISME Journal (2021)
-
Spatial patterns in phage-Rhizobium coevolutionary interactions across regions of common bean domestication
The ISME Journal (2021)
-
Phage mobility is a core determinant of phage–bacteria coexistence in biofilms
The ISME Journal (2018)
-
Bacteriophages and its applications: an overview
Folia Microbiologica (2017)