Abstract
The development of DNA sequencing methods for characterizing microbial communities has evolved rapidly over the past decades. To evaluate more traditional, as well as newer methodologies for DNA library preparation and sequencing, we compared fosmid, short-insert shotgun and 454 pyrosequencing libraries prepared from the same metagenomic DNA samples. GC content was elevated in all fosmid libraries, compared with shotgun and 454 libraries. Taxonomic composition of the different libraries suggested that this was caused by a relative underrepresentation of dominant taxonomic groups with low GC content, notably Prochlorales and the SAR11 cluster, in fosmid libraries. While these abundant taxa had a large impact on library representation, we also observed a positive correlation between taxon GC content and fosmid library representation in other low-GC taxa, suggesting a general trend. Analysis of gene category representation in different libraries indicated that the functional composition of a library was largely a reflection of its taxonomic composition, and no additional systematic biases against particular functional categories were detected at the level of sequencing depth in our samples. Another important but less predictable factor influencing the apparent taxonomic and functional library composition was the read length afforded by the different sequencing technologies. Our comparisons and analyses provide a detailed perspective on the influence of library type on the recovery of microbial taxa in metagenomic libraries and underscore the different uses and utilities of more traditional, as well as contemporary ‘next-generation’ DNA library construction and sequencing technologies for exploring the genomics of the natural microbial world.
Similar content being viewed by others
Introduction
Nucleic acid-based cultivation-independent surveys (CIS) of natural microbial assemblages have revolutionized the practice, knowledge base and theory in microbial evolution and ecology (Woese, 2004; Pace, 1997, 2009). CIS strategies, first conceived by Pace and collaborators over 25 years ago (Pace et al., 1985; Olsen et al., 1986), were inspired by the seminal microbial evolutionary studies of Woese (1987). Pace and colleagues realized that the ability to infer identity and evolutionary relationship from macromolecular sequence data sidestepped the necessity of cultivating microbes in order to identify them. At the time that CIS approaches were first conducted (initially by directly isolating and sequencing 5S ribosomal RNA (rRNA); Stahl et al., 1984), the polymerase chain reaction (Saiki et al., 1988) was not in widespread use. So Pace and collaborators extended CIS strategies to leverage the cloning and sequencing of large DNA fragments extracted from mixed microbial communities (Pace et al., 1985; Olsen et al., 1986; Schmidt et al., 1991), an approach later referred to as ‘metagenomics’ (Rondon et al., 2000). The basic method involved extracting and purifying DNA from natural microbial biomass, cloning of large DNA fragments into phage vectors and the subsequent identification of clones containing phylogenetically informative rRNA sequences. Subsequent nucleic acid sequencing and phylogenetic analysis of rRNA genes allowed the identification of microbes found in the original mixed population. One of the first successful applications of the approach used lambda phage clone libraries prepared from marine bacterioplankton community DNA to successfully identify major bacterial groups that predominate in seawater (Schmidt et al., 1991). This early study pointed out that besides recovering rRNA sequences, genomic libraries prepared from microbial community DNA also provided access to other sequences and genes of interest (Schmidt et al., 1991).
During the 1990s, microbial CIS methods and applications were further developed, in tandem with the improvement of DNA sequencing technologies. Because of their ease of application and high throughput, primer-driven rRNA PCR amplification techniques initially became the method of choice for early rRNA-based evolutionary and environmental studies (Medlin et al., 1988; Giovannoni et al., 1990). CIS approaches in environmental microbiology gradually extended beyond PCR-based studies that were focused on single-gene phylogenetic markers, to the sequencing of entire cloned DNA fragments, combining bacterial artificial chromosome (BAC) vectors (Stein et al., 1996; Béjà et al., 2000a, 2000b) and small-insert shotgun sequencing approaches and employing improved high-throughput Sanger-based dideoxynucleotide terminator sequencing (Venter et al., 2004). More recently, cloning-independent ‘next-generation’ sequencing technologies such as pyrosequencing (Edwards et al., 2006) have enabled more economical, technically streamlined approaches, which have ‘democratized’ the accessibility of large-scale DNA sequencing. As a result, ‘next-generation’ CIS approaches are currently being applied to a vast array of naturally occurring microbial assemblages.
In part because of the rapid development of different technologies, there have been relatively few systematic comparisons of the efficacy, advantages and disadvantages of different CIS approaches for characterizing microbial communities. Shotgun sequencing techniques originally had provided the greatest depth of coverage and throughput, but genetic heterogeneity within natural populations render assembly of large contigs difficult at best, in all but the simplest populations (Tyson et al., 2004; DeLong, 2005; Tringe et al., 2005). Longer DNA sequence contigs approaching 100 kb or more can be accessed in BAC or fosmid libraries, but the quantitative recovery of microbial assemblage DNA from such large-insert libraries is not well characterized. ‘Next-generation’ sequencing technologies, such as pyrosequencing and Illumina, avoid potential cloning biases, are affordable and rapid, and provide very deep sequence coverage. Nevertheless, ‘next-generation’ sequencing technologies typically have greater error rates and shorter read lengths than Sanger-based sequencing methods, and potential for systematic biases as well (MacLean et al., 2009).
To gain a better perspective on the advantages, disadvantages and potential biases of different libraries and sequencing technologies for metagenomic analysis, we compared data sets from fosmid, shotgun and 454 sequencing libraries that were prepared from the same metagenomic DNA. Previous studies reported an apparent underrepresentation of the SAR11 cluster of the Alphaproteobacteria in BAC and fosmid libraries (Pham et al., 2008; Feingersch and Béjà, 2009; Temperton et al., 2009). Temperton et al. (2009) proposed that the low GC content of Candidatus Pelagibacter rendered its DNA more susceptible to fracturing, thereby biasing its inclusion in libraries requiring large insert sizes. Feingersch and Béjà (2009) on the other hand speculated that the expression of certain genes, in particular ribosomal proteins, may prove toxic to the Escherichia coli host and that BAC libraries are more affected than shotgun libraries, because their larger inserts are more likely to contain at least one such toxic sequence. Subsequently, Ghai et al. (2010) reported an apparent GC bias and under representation of Pelagibacter and Prochlorococcus clones in picoplankton libraries from a Mediterranean deep-chlorophyll maximum layer (Ghai et al., 2010). To help clarify some of these issues, we report here a more detailed analysis of taxon representation and potential biases associated with multiple sets of deeply sequenced fosmid, shotgun and 454 sequencing libraries, each set prepared from the same sample of open-ocean picoplankton DNA.
Materials and methods
Hot data sets
The bacterioplankton samples (size fraction 0.22–1.6 μm) for the preparation of the libraries used in this comparison were collected as described previously (Frias-Lopez et al., 2008; Shi et al., 2009, 2010) as part of the Hawaii Ocean Times Series (HOT) Project (Karl and Lukas, 1996). The HOT 179 collection consists of fosmid, shotgun and 454 libraries from samples collected in March 2006 at four different depths—25 m, 75 m, 125 m and 500 m. Samples from the HOT 186 cruise were obtained in October 2006 at 25 m, 75 m, 110 m and 500 m depth. For this collection shotgun libraries are available for 25 m and 75 m, and fosmid as well as 454 sequences for all depths. Table 1 summarizes the properties of the different data sets. All libraries and their construction have been described previously (DeLong et al., 2006; Frias-Lopez et al., 2008; Martinez et al., 2010; Shi et al., 2010). The fosmid and short-insert shotgun libraries were sequenced at the Joint Genome Institute (JGI, Walnut Creek, CA, USA; protocols can be found at http://www.jgi.doe.gov/sequencing/protocols/prots_production.html). The pyrosequencing libraries were prepared and sequenced according to standard protocols recommended by 454 Life Sciences (Branford, CT, USA) for the respective technology (GS 20 for HOT 179, and FLX for HOT 186 samples) as previously described (Frias-Lopez et al., 2008). Reads with fewer than 60 unambiguous nucleotides were not included in the analysis. All nucleotide sequences are available from public databases, and accession numbers as well as cloning vectors are listed in Supplementary Table 2.
Clustering
Highly similar and overlapping sequences in each library were clustered using the ‘EST’ (expressed sequence tags) program of the CD-HIT package (version 4.0; http://weizhong-lab.ucsd.edu/cd-hit/) with parameters -n 10 -b 20 -c 0.98 -r 1, applying an identity threshold of 98% while comparing each sequence and its reverse complement to all other sequences. Representative sequences were used for further analysis and any counts were subsequently adjusted using the number of reads that each sequence represented after clustering.
Blast searches
Nucleotide BLAST analysis was performed using the NCBI blastn program (Altschul et al., 1997, version 2.2.16) against a comprehensive, nonredundant collection of microbial genome sequences compiled in July 2010 from the Gordon and Betty Moore Foundation Microbial Genome Sequencing Project (http://camera.calit2.net/microgenome/), as well as from the Bacteria, Viruses, Plasmids, Protozoa and Fungi subdirectories of the NCBI’s genomes repository (ftp://ftp.ncbi.nih.gov/genomes/). This database, which contains 13 768 sequences and 5.1 billion nt, gave essentially the same results for the samples in question as using the NCBI nt database (data not shown), while reducing the search time by an order of magnitude. The blastn program was used with parameters for filtering as recommended for processing with MEGAN (DH Huson et al., Tübingen University, Germany): DUST filter, with mask for lookup table only: -F ’D; m’.
For comparison, blastx was used with a protein database assembled from the same sources described above for the nucleotide sequences, which contained 3.6 million peptides with a total of 1.2 billion residues. For functional analysis we used blastx against a KEGG database (Kanehisa and Goto, 2000; Kanehisa, 2006). All blastx analyses were performed with genetic code table 11 (bacterial sequences) and filtering using the ‘SEG’ algorithm, masking for lookup table only. In all BLAST searches GI numbers were included (-I T) and an E-value cutoff of 10−3 was used.
Taxonomic classification
The MEGAN program (version 3.9; Huson et al., 2007) was used to assign BLAST hits to taxa of the NCBI taxonomy with the following parameters for the ‘LCA’ (lowest common ancestor) algorithm: min support 1 (as sequences were already clustered), min score 35 (as recommended for 454 sequences), top percent 10, and win score 0. Where possible (fosmid ends and shotgun data sets), the ‘paired reads’ setting was used. Read assignments were exported, yielding a list of reads associated with each taxon.
Calculation of bias in distribution of taxa between data sets
For each sequence library, read assignments to taxa by MEGAN were combined with sequence data, as well as clustering information and further processed using a program written in Perl to create tables containing the counts, aggregate counts (including all child taxa) and GC content statistics (mean and s.d. for sequences assigned to a taxon; both individual and aggregate). In addition, an alternate set of aggregate counts and GC statistics was calculated, which excludes the SAR11 cluster (NCBI taxon ID 54 526) and the Prochlorales (NCBI taxon ID 1212), as well as all their child taxa. The purpose of this alternate set was to allow for a more accurate comparison of other groups after removing the numerically dominant taxa that show a high bias in their distribution between data sets.
As a measure for the bias toward one data set or another, sets were compared pairwise, by calculating the base-2 logarithm of the ratio of the relative abundance in one set over the other, for example,  a value of −1 in this context means that the given taxon is twice as frequent in the 454 data set as it is in the fosmid library, and +2 signifies a fourfold bias in favor of the fosmid set, whereas 0 indicates equal distribution. Calculations of the relative abundance of taxa in data sets and of the bias in sequence library comparisons were performed using the R statistical computing software (R Development Core Team, 2009).
a value of −1 in this context means that the given taxon is twice as frequent in the 454 data set as it is in the fosmid library, and +2 signifies a fourfold bias in favor of the fosmid set, whereas 0 indicates equal distribution. Calculations of the relative abundance of taxa in data sets and of the bias in sequence library comparisons were performed using the R statistical computing software (R Development Core Team, 2009).
Functional classification
For each library the BLAST results against the KEGG database with an E-value cut-off of 10−3 were parsed using a Perl script to assign counts to each ortholog. Only hits that could be assigned to specific KEGG gene/protein ortholog groups with K-numbers were considered in this analysis. The counts for each of the three libraries were analyzed separately for each sample in HOT 179 using version 1.6.0 of the baySeq package (Hardcastle, 2011; Hardcastle and Kelly, 2010) in version 2.13.1 of the R statistics software (R Development Core Team, 2009).
The baySeq algorithm estimates for each KEGG ortholog the probabilities of all possible models for the parameters of the Poisson distributions that apply to each library—all parameters are the same (AAA), all are different (ABC) or one is different and the others are the same (ABB). As the last possibility exists for all three libraries, five models are compared in our case. As all libraries were prepared from the same DNA, unbiased distribution of a gene would favor model AAA. The probability that a library is positively or negatively biased with respect to a specific ortholog is the sum of the models ABB and ABC. To consider any effect in a library–ortholog combination significant, we impose the following criteria: (1) The ortholog has to have the same type of bias (positive or negative) when compared with both other libraries, otherwise this library would be considered ‘neutral’. (2) All samples with an estimated probability of being biased of 0.5 or greater have to be biased in the same direction. (3) To reduce the rate of false discovery, we only considered orthologs that show probabilities of ⩾0.5 in at least three of the four samples (25, 75, 125 and 500 m), or of ⩾0.9 in two or more samples when analyzing a specific library.
Results and Discussion
Taxonomic composition of the data sets
To evaluate any differences in taxon recovery between fosmid, shotgun and 454 libraries, we used blastn and MEGAN to assign each read to a taxon. Depending on the sample and library type, between 10 and 78% of the reads had BLAST hits that could be successfully assigned by MEGAN (Table 1). Other commonly used methods for taxonomic classification employ either rRNA or protein databases (the latter with blastx). As discussed by Shi et al., 2010, classification of taxa by protein-coding sequences differs from rRNA-based analysis in some details, while general distributional trends between them are similar. For the purpose of the analysis reported here, we chose not to perform rRNA-based comparisons, as our goal was to compare DNA recovery in the different library types on the broadest possible basis, rather than restricting the analyses to a small fraction of the reads.
Use of blastx against a peptide database increased the identification rate to 26–91% in a largely read-length-dependent manner, with the short 454 GS 20 sequences recovering fewer matches than the longer shotgun and fosmid reads (Table 1). Regardless of the BLAST method, the short-insert shotgun libraries had the greatest number of database matches, while the fosmid and 454 libraries contained a greater proportion of unidentified sequences (Table 1), for reasons discussed below. The results from nucleotide and peptide BLAST-based analyses differ in absolute numbers, but show the same general trends (see Figure 1 vs Supplementary Figure 1, Supplementary Figure 2 vs 3, Supplementary Tables 3–4 vs 5–6 and 7–8 vs 9–10, Figure 2 vs Supplementary Figure 5, Figures 3a–c vs Supplementary Figures 9c–e). We focus the rest of the discussion on the nucleotide results, as they are a more direct measure of sequence identity shared between different libraries.
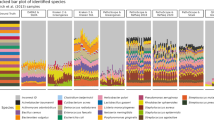
Taxonomic comparison of libraries in HOT 179, analyzed using blastn. A representative subset of taxa is shown and their indentations reflect the hierarchical rank in the NCBI taxonomy. The number of reads assigned to each taxon, including its child taxa, is displayed as percentage of all successfully assigned reads in the library.
Distribution of the bias measure in pairwise comparisons of library types, using blastn analysis results. Positive and negative infinity of the bias reflect the absence of some taxa in one of the libraries. The colors of the histogram bars are indicative of the library types being compared. The y axis shows the percentage of reads assigned to taxa falling into the respective bias interval, averaged over both libraries. Data is shown for four samples, HOT-179 25 m (a, b), 75 m (c, d) and 500 m (e, f), as well as HOT 186-75 m (g, h). Panels a, c, e and g show the results of calculating the bias using all taxa, whereas panels b, d, f and h omit the Prochlorales and the SAR11 cluster, as described in Materials and methods.
GC contents plotted versus bias for pairwise library comparisons using blastn. Panels (a–c) show the combined photic-zone samples and (d–f) the 500-m sample of HOT 179. Each taxon is represented by two symbols, one for the GC content in each library. Taxa with less than five reads in each library are only shown if they are not represented at all in one library (infinite bias measure), where their expected representation based on their percentage in the other library would be at least two reads.
The representation of the major taxonomic groups in the identified reads of the samples is shown in Figure 1 (nucleotide BLAST of HOT 179), Supplementary Figures 1–3 (HOT 186 and peptide BLAST of HOT 179) and Supplementary Tables 3–10. All samples were dominated by Bacteria, which accounted for 90% or more of the assigned reads in most samples. While eukaryotic and viral sequences were present, their counts were comparatively low, as expected because of the sample collection and filtration methods used. Archaea represented <1% of most libraries, except for the 500-m samples, where they approached 10% or more of all taxa identified.
The three different library construction methods each provided a different quantitative representation of the taxonomic composition in the samples. In the photic-zone cyanobacteria, specifically the Prochlorales, comprised typically more than half of the identified bacterial sequences in the 454 libraries, whereas their representation was at least twofold reduced in the fosmid libraries. The only exception to this pattern was the sample HOT 186 from 110 m, where the situation was reversed and 67% of the fosmid library and only 42% of the 454 library were associated with the order Prochlorales (see Supplementary Table 8 and Supplementary Figure 2). The one other prevalent taxonomic group that was consistently underrepresented by a large factor in the fosmid libraries, relative to the shotgun and 454 libraries, was the SAR11 cluster, which comprised between 20 and 30% of assigned reads in most 454 libraries, but only between 0.6 and 3.5% in fosmid libraries.
The reduction in the percentage of reads in the numerically dominant taxa in the fosmid libraries predicts an expected general increase in fractions from other taxa. The reduced representation of the most abundant taxa in fosmid libraries may also explain the higher percentage of reads without BLAST hits in samples dominated by the Prochlorales, as a shortfall of the taxa well-represented in the sequence databases leads to a relative increase in other taxa that are less likely to have a BLAST match. The lower number of BLAST hits in the 454 libraries, on the other hand, is most likely a result of the shorter read length, which provides fewer opportunities for partial BLAST matches than longer sequences.
In contrast, taxa that were overrepresented in fosmid libraries in comparison with shotgun and 454 libraries included the Gammaproteobacteria, the Deltaproteobacteria, the Rhodobacterales and in some libraries the Rhizobiales (Figure 1, Supplementary Figures 1–3, Supplementary Tables 3–10). For the Rhodobacterales this was particularly the case in the 25-m and 75-m samples of HOT 186 (Supplementary Figures 2 and 3, Supplementary Tables 7 and 9). In these same samples the high representation of the Gammaproteobacteria in fosmid libraries was a result of increased numbers of sequences from Alteromonadales, but the latter are not dominant among the Gammaproteobacteria in the other samples.
In the 500-m samples overrepresentation of any taxa in the fosmid libraries was either greatly diminished or absent, consistent with a decreased dominance of underrepresented groups like the Prochlorales. These samples contained a greater fraction of unidentified reads, regardless of library type, as genome sequences from this depth are sparsely represented in the databases. Almost half the identified sequences in the mesopelagic zone at the HOT station mapped to Alpha- and Gammaproteobacteria, particularly the SAR11 cluster at 20 to 25% sequence representation. Actinobacteria comprised 3 to 8% of the assigned reads at this depth, compared with <1% in most shotgun and 454 libraries and <4% in most fosmid libraries from the photic zone. Perhaps surprisingly, Cyanobacteria-like sequences were still detected at 500 m, especially in HOT 186, albeit at very low abundance. Apparent archaeal sequence representation exceeded 1% only in the mesopelagic samples. At this depth most archaeal reads were assigned to the Thaumarchaeota, which—unlike the Euryarchaeota—were underrepresented in fosmid and overrepresented in shotgun libraries, compared with 454.
Differential representation of taxa in sequencing libraries
To further investigate the over- or underrepresentation of individual taxa in each type of library, we calculated the relative representation of each taxon in pairwise comparisons between libraries prepared from the same DNA sample. The bias for any given taxon is calculated as the base-2 logarithm of the ratio of the relative abundance in one library type over the other (Materials and methods). As unbiased library construction should theoretically sample populations stochastically, we would ideally expect a symmetrical distribution of the bias measure around zero. The larger the number of reads that are assigned to a given taxon, the more likely is it that the bias measure accurately represents the actual sampling bias.
In histograms showing the distribution of the bias in different samples, each taxon was weighted according to its representation in both libraries to reduce the impact of rare taxa, which are more likely to show large random deviations (Figure 2, Supplementary Figures 4 and 5). In addition we excluded reads that did not have BLAST hits or those with database matches that were not assigned to taxa by MEGAN (see Table 1; while the former category comprised the majority in most data sets, the latter applied to <0.1% of the reads in most samples and was always <0.3%). Comparison of shotgun versus 454 libraries (blue bars, Figures 2a, c) typically showed a distribution of the bias around zero, whereas the comparisons involving fosmid libraries (red and yellow bars, Figures 2a, c) exhibited bimodal distributions when all taxa were included. This was due to a small number of dominant taxa that were underrepresented in the fosmid libraries (negative bias in Figure 2), which resulted in a relative overrepresentation and therefore positive bias of most other taxa.
As the Candidatus Pelagibacter and Prochlorococcus strains were underrepresented in fosmid libraries, but represent the most ubiquitous species, particularly in the photic zone (25–125-m samples, see Figure 1 and Supplementary Tables 3–10), we recalculated the bias distribution excluding the SAR11 cluster and the Prochlorales to examine their effect. For most samples this resulted in an unimodal distribution of the bias around zero for all comparisons, confirming that most of the observed bias was caused by the abundant SAR11 and Prochlorales (Figures 2b, d and f). While other taxa were represented unevenly as well, they were not sufficiently prevalent to cause large shifts in the distribution.
A notable exception to this was the 75-m sample of HOT 186, where the bimodal distribution was also evident in the shotgun/454 comparison in addition to those involving fosmid libraries (Figure 2g). The shape of this distribution persisted, particularly in the comparisons including shotgun, when SAR11 and Prochlorales were excluded from the analysis (Figure 2h), suggesting the presence of other taxa with uneven representation across all the libraries. Closer examination revealed that comparatively large numbers of reads were assigned to ‘Bacteria’ and ‘cellular organisms’ by MEGAN. Combined, these indeterminate groups accounted for 39% of assigned reads in the shotgun library, compared with 4% and 7%, respectively, in the 454 and fosmid libraries. When calculating the bias without SAR11 and Prochlorales, ‘Bacteria’ and ‘cellular organisms’ combined favored shotgun over fosmids with a bias of 3.06 and 454 over fosmids with 1.20. The bias direction in the 454/fosmids comparison changed after removal of Prochlorales and SAR11, because this increased the relative weight of the indeterminate taxa in the 454 library significantly, while affecting the fosmid library only marginally. The reads assigned to ‘Bacteria’ and ‘cellular organisms’ typically had low-scoring blastn hits of comparable magnitude (within the 10% of the top hit) to species in different phyla or even superkingdoms, indicating that they originated from uncharacterized taxa, which were not well represented in the sequence databases, but had sufficiently conserved features to allow significant matches in a blastn search. Comparison with blastx results for the same reads from the shotgun library showed that 33% could not be assigned to any more specific taxon, whereas 7% were classified as Proteobacteria, 7% as Enterobacteriaceae, 4% as Gammaproteobacteria, 4% as Alphaproteobacteria, 4% as Rhodobacteraceae, and the rest as various other taxa. The GC content for bacteria/cellular organisms was 49%/45% in fosmids, 41%/50% in shotgun, and 37%/35% in 454, which may explain why these taxa were more highly represented in 454 than fosmid libraries. The GC content cannot account for the even higher prevalence of ‘Bacteria’ and ‘cellular organisms’ in the shotgun library, however, which may be due to its greater average read length, which was more than four times that of the 454 library and almost 2.5 times that of the fosmid library (Table 1).
GC content and bias
Temperton et al. (2009) previously suggested that GC content may have a substantial impact on taxon representation in metagenomic libraries. To further test for a potential relationship between the taxon GC content and library representation, we calculated the average GC content for each taxon in each library (weighting each sequence by length) and plotted the results against the apparent bias (see Figure 3 and Supplementary Figures 6–9). While the taxa in the shotgun/454 comparison (Figures 3c and f) were—with the exception of a few outliers—relatively tightly grouped around zero bias, irrespective of their GC content, this did not hold true for the comparisons involving fosmid libraries (Figures 3a, b). Especially for the photic zone (Figures 3a–c), there appeared to be a positive correlation between low GC content and exclusion from the fosmid library, whereas most taxa with higher GC contents were tightly clustered and showed no obvious relationship between GC and bias. While most taxa appeared to have a positive bias in these comparisons, this is an artifact of the high abundance of the Prochlorales, which are negatively biased and thus shift the remaining taxa in the other direction. The 500-m samples (Figures 3d–f) did not have as many low-GC taxa, but those that did exist also showed a correlation of GC content and bias. In addition, the GC content of low-GC taxa in the fosmid libraries was almost always several percentage points higher than that found in corresponding taxa in shotgun or 454 libraries of the same sample. This trend, however, only held for a GC content of < ∼45%. For example, the average difference between GC contents of a taxon in fosmid and 454 libraries (GCfosmids − GC454) for the combined photic zone of HOT 179 (Figure 3a) was −1.6% for taxa with a GC content >45%, but 6.1% for taxa having <45% GC (for this classification we used the lower of the two library GC contents).
To test the effect of the read length and the resulting biases in taxon representation, we randomly split the fosmid and shotgun sequences of HOT 179 125 m into shorter fragments with an average length of 108.8nt, which is equivalent to the mean length of the 454 sequences. Supplementary Figure 7 shows that eliminating length differences does not significantly affect the general trend in the relationship between the GC content and bias.
These observations support the previous suggestion of Temperton et al. (2009) and Ghai et al. (2010), that low-GC taxa in general are less well represented in fosmid libraries, compared with small-insert shotgun clone libraries or 454 sequencing libraries, and that low-GC content itself may be the responsible factor for poor representation in the fosmid libraries. While the GC content does appear to be responsible for much of the observed bias, other factors such as toxic gene products, or chemically modified nucleotide content in the DNA, may also have a role.
Interestingly, the Candidatus Pelagibacter species appear to be much more strongly biased against the fosmid libraries in most samples than other taxa with similar GC content. This suggests additional causes for the underrepresentation, such as the toxicity suggested by Feingersch and Béjà (2009), or phosphorothioation of the DNA, which can accelerate its degradation (Wang et al., 2007, 2011). Homologs of genes responsible for the latter have been found in several marine metagenomes and in at least one Candidatus Pelagibacter strain, but are not common to all strains (He et al., 2007; Wang et al., 2011). DNA degradation resulting from phosphorothioation of DNA—similar to low-GC—might affect libraries requiring larger DNA fragments more than those with short inserts.
Functional analysis
To assess whether the library type significantly affects estimates of functional gene categories, we analyzed the assignment of KEGG orthologs to the three library types. An empirical Bayesian analysis (Hardcastle and Kelly, 2010) was performed independently for each depth in HOT 179. KEGG orthologs were classified as significantly biased or not by comparing the probability and direction of bias for or against a library type between different samples (Materials and methods). This approach minimizes false positives, while taking into account that only very few, if any, orthologs showed a consistent bias with probability exceeding 0.5 at all four depth (two for the fosmid library, none for shotgun, six for 454). All gene categories with significant biases detected by this analysis are listed in Supplementary Tables 11–15.
Of the 7813 KEGG orthologs identified in all the HOT 179 samples, only four were biased in favor of the shotgun libraries according to our criteria, and none against. One of them (K04744, unclassified ion channel) was also biased against the 454 libraries, indicating a read length effect, while the others showed no significant bias in other libraries. The fosmid libraries showed a significant bias against 17 orthologs. Four of these have functions in DNA replication and repair, three in photosynthesis, two in carbohydrate metabolism and the others fall into various mostly metabolic categories. The combined total of the reads for these 17 orthologs is 2.3% of all assigned reads in the 454 libraries and 1.7% in the shotgun libraries, compared with 0.6% in the fosmid libraries across all depths. In contrast we found 34 KEGG orthologs biased in favor of the fosmid libraries, comprising 4.7% of assigned fosmid reads, compared with 1.5% and 0.4% in shotgun and 454, respectively. These comparatively small percentages make it unlikely that a functional bias is solely responsible for the observed large shift in taxonomic distribution. It is more plausible that the functional composition largely mirrors the taxonomic composition; for example, the reduction of any photosynthesis-associated orthologs in the fosmid libraries can be explained by the reduction in the cyanobacteria. Examination of the taxa that contribute reads to the biased KEGG orthologs supports this hypothesis—categories underrepresented in the fosmid libraries map largely to the most prevalent underrepresented species, Prochlorococcus marinus, whereas overrepresented orthologs come from a broad range of taxa.
Another reason for biased representation of a functional category in one library type over another are biased BLAST results caused by the varying length of the sequences available in the different library types. This is evident in the comparison of libraries with different read lengths, such as 454 versus shotgun. In fact, many of the functions biased in favor of the fosmid libraries are likely a result of the shorter read length in the 454 sequences. This is corroborated by the observation that 11 of these 34 orthologs were significantly reduced in the 454 libraries. The latter showed a bias against 253 orthologs, which in the shotgun libraries amount to 7.2%, suggesting that read length has an important role in the perceived functional composition. Short reads are more likely to find a short but close match among the taxa and functions that are well represented in the sequence databases, whereas longer reads may bridge domains and can therefore accumulate a sufficiently high score even in less common proteins, where an exact match is less likely. On the other hand, 41 orthologs (5.7% of the assigned shotgun reads) showed a positive bias toward 454. Several of those had functions in photosynthesis, indicating that for at least some orthologs the positive bias in 454 may be a reflection of the lack of the corresponding taxa in the fosmid libraries, even though only K03798 (a peptidase) meets our criteria for significance. This is supported by the observation that the majority of overrepresented orthologs could be assigned to either cyanobacteria or Candidatus Pelagibacter using blastx results against the microbial protein database. In contrast, the underrepresented categories originate from many different taxa, including SAR11 and cyanobacteria.
The third possible explanation for differential representation of a gene category across library types is toxicity to the cloning host E. coli (Sorek et al., 2007), which might be expected to exert the greatest influence on fosmids or BAC libraries, which contain large inserts. Our library sizes were too small to permit meaningful simultaneous analysis of both functional and taxonomic representation and so we cannot identify toxic proteins with any degree of confidence. Although toxic gene products may have some influence, other factors, including read length and taxonomic composition appear to have a larger effect on KEGG profile differences observed between the different sequencing libraries.
Conclusions
The use of different library types and sequencing methods for metagenomics is largely a matter of historical development, with respect to both technological innovation and cost effectiveness. Different metagenomic libraries and sequencing technologies have different optimal usages and applications. For example, large-insert BAC and fosmid libraries allow for the stable recovery of large DNA fragments ranging from 35 to >100 kb in size. This provides access to large contigs that otherwise would be difficult, if not impossible, to assemble—especially for rarer taxa—from DNA sequences of complex microbial community assemblages (Stein et al., 1996; Béjà et al., 2000a, 2000b). In addition, the larger fragments can harbor entire multigene complexes, operons and metabolic pathways, that can be functionally expressed and studied from the perspective of biochemistry and metabolism (Martinez et al., 2007, 2010). BAC and fosmid libraries are useful archival resources as well, as new targets and screening approaches are developed. As demonstrated conclusively in this study, however, these library types have the potential to yield biased quantitative representations of resident microbial taxa, particularly those containing genomes with low GC content or DNA modifications leading to increased degradation.
Over the past five years, clone-free ‘next-generation’ DNA sequencing technologies have largely supplanted clone libraries and Sanger-based sequencing for environmental DNA sequencing surveys. High-throughput technologies such as 454 pyrosequencing and Illumina generate shorter reads, but offer lower cost, greater sequencing depth and—provided that read-duplication and sequencing errors are dealt with appropriately—appear relatively unbiased. They can therefore prove more useful for a quantitative analysis of community composition, but individual sequences may carry less information, due to the generally shorter average read lengths compared with Sanger sequences. This too, however, is rapidly changing, as pyrosequencing read lengths now approach that of Sanger-based sequencing technologies. The combination of old and new technologies—large-insert libraries for archival purposes and functional gene and metabolic pathway studies, and next-generation sequencing techniques for surveys—provide useful and complementary tools for probing the diversity, structure and function in the natural microbial world.
Change history
16 October 2012
This article has been corrected since Advance Online Publication and a corrigendum is also printed in this issue.
References
Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W et al. (1997). Gapped blast and psi-blast: a new generation of protein database search programs. Nucleic Acids Res 25: 3389–3402.
Béjà O, Aravind L, Koonin EV, Suzuki MT, Hadd A, Nguyen LP et al. (2000a). Bacterial rhodopsin: evidence for a new type of phototrophy in the sea. Science 289: 1902–1906.
Béjà O, Suzuki MT, Koonin EV, Aravind L, Hadd A, Nguyen LP et al. (2000b). Construction and analysis of bacterial artificial chromosome libraries from a marine microbial assemblage. Environ Microbiol 2: 516–529.
DeLong EF . (2005). Microbial community genomics in the ocean. Nat Rev Microbiol 3: 459–469.
DeLong EF, Preston CM, Mincer T, Rich V, Hallam SJ, Frigaard N et al. (2006). Community genomics among stratified microbial assemblages in the ocean’s interior. Science 311: 496–503.
Edwards R, Rodriguez-Brito B, Wegley L, Haynes M, Breitbart M, Peterson D et al. (2006). Using pyrosequencing to shed light on deep mine microbial ecology. BMC Genomics 7: 57.
Feingersch R, Béjà O . (2009). Bias in assessments of marine SAR11 biodiversity in environmental fosmid and BAC libraries? ISME J 3: 1117–1119.
Frias-Lopez J, Shi Y, Tyson GW, Coleman ML, Schuster SC, Chisholm SW et al. (2008). Microbial community gene expression in ocean surface waters. Proc Natl Acad Sci USA 105: 3805–3810.
Ghai R, Martin-Cuadrado AB, Molto AG, Heredia IG, Cabrera R, Martin J, Verdú M, Deschamps P, Moreira D, López-García P, Mira A, Rodriguez-Valera F . (2010). Metagenome of the Mediterranean deep chlorophyll maximum studied by direct and fosmid library 454 pyrosequencing. ISME J 4: 1154–1166.
Giovannoni SJ, Britschgi TB, Moyer CL, Field KG . (1990). Genetic diversity in Sargasso Sea bacterioplankton. Nature 345: 60–63.
Hardcastle TJ . (2011). baySeq: empirical Bayesian analysis of patterns of differential expression in count data. R package version 1.6.0.
Hardcastle TJ, Kelly KA . (2010). baySeq: empirical Bayesian methods for identifying differential expression in sequence count data. BMC Bioinformatics 11: 422.
He X, Ou H, Yu Q, Zhou X, Wu J, Liang J et al. (2007). Analysis of a genomic island housing genes for DNA S-modification system in Streptomyces lividans 66 and its counterparts in other distantly related bacteria. Mol Microbiol 65: 1034–1048.
Huson DH, Auch AF, Qi J, Schuster SC . (2007). MEGAN analysis of metagenomic data. Genome Res 17: 377–386.
Kanehisa M . (2006). From genomics to chemical genomics: new developments in KEGG. Nucleic Acids Res 34: D354–D357.
Kanehisa M, Goto S . (2000). KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res 28: 27–30.
Karl DM, Lukas R . (1996). The Hawaii Ocean Time-series (HOT) program: Background, rationale and field implementation. Deep-Sea Res Pt II 43: 129–156.
MacLean D, Jones JDG, Studholme DJ . (2009). Application of ‘next-generation’ sequencing technologies to microbial genetics. Nat Rev Microbiol 7: 287–296.
Martinez A, Bradley AS, Waldbauer JR, Summons RE, DeLong EF . (2007). Proteorhodopsin photosystem gene expression enables photophosphorylation in a heterologous host. Proc Natl Acad Sci USA 104: 5590–5595.
Martinez A, Tyson GW, DeLong EF . (2010). Widespread known and novel phosphonate utilization pathways in marine bacteria revealed by functional screening and metagenomic analyses. Environ Microbiol 12: 222–238.
Medlin L, Elwood HJ, Stickel S, Sogin ML . (1988). The characterization of enzymatically amplified eukaryotic 16S-like rRNA-coding regions. Gene 71: 491–499.
Olsen GJ, Lane DJ, Giovannoni SJ, Pace NR, Stahl DA . (1986). Microbial ecology and evolution: A ribosomal RNA approach. Annu Rev Microbiol 40: 337–365.
Pace NR . (1997). A molecular view of microbial diversity and the biosphere. Science 276: 734–740.
Pace NR . (2009). Mapping the tree of life: Progress and prospects. Microbiol Mol Biol Rev 73: 565–576.
Pace NR, Stahl DA, Olsen GJ, LD J . (1985). Analyzing natural microbial populations by rRNA sequences. ASM News 51: 4–12.
Pham VD, Konstantinidis KT, Palden T, DeLong EF . (2008). Phylogenetic analyses of ribosomal DNA-containing bacterioplankton genome fragments from a 4000 m vertical profile in the North Pacific Subtropical Gyre. Environ Microbiol 10: 2313–2330.
R Development Core Team (2009) R: A Language and Environment for Statistical Computing R Foundation for Statistical Computing: Vienna, Austria.
Rondon MR, August PR, Bettermann AD, Brady SF, Grossman TH, Liles MR et al. (2000). Cloning the soil metagenome: a strategy for accessing the genetic and functional diversity of uncultured microorganisms. Appl Environ Microbiol 66: 2541–2547.
Saiki RK, Gelfand DH, Stoffel S, Scharf SJ, Higuchi R, Horn GT et al. (1988). Primer-directed enzymatic amplification of DNA with a thermostable DNA polymerase. Science 239: 487–491.
Schmidt TM, DeLong EF, Pace NR . (1991). Analysis of a marine picoplankton community by 16 S rRNA gene cloning and sequencing. J Bacteriol 173: 4371–4378.
Shi Y, Tyson GW, DeLong EF . (2009). Metatranscriptomics reveals unique microbial small RNAs in the ocean’s water column. Nature 459: 266–269.
Shi Y, Tyson GW, Eppley JM, DeLong EF . (2010). Integrated metatranscriptomic and metagenomic analyses of stratified microbial assemblages in the open ocean. ISME J 5: 999–1013.
Sorek R, Zhu Y, Creevey CJ, Francino MP, Bork P, Rubin EM . (2007). Genome-wide experimental determination of barriers to horizontal gene transfer. Science 318: 1449–1452.
Stahl DA, Lane DJ, Olsen GJ, Pace NR . (1984). The analysis of hydrothermal vent-associated symbionts by ribosomal RNA sequences. Science 224: 409–411.
Stein JL, Marsh TL, Wu KY, Shizuya H, DeLong EF . (1996). Characterization of uncultivated prokaryotes: isolation and analysis of a 40-kilobase-pair genome fragment from a planktonic marine archaeon. J Bacteriol 178: 591–599.
Temperton B, Field D, Oliver A, Tiwari B, Mühling M, Joint I et al. (2009). Bias in assessments of marine microbial biodiversity in fosmid libraries as evaluated by pyrosequencing. ISME J 3: 792–796.
Tringe SG, von Mering C, Kobayashi A, Salamov AA, Chen K, Chang HW et al. (2005). Comparative metagenomics of microbial communities. Science 308: 554–557.
Tyson GW, Chapman J, Hugenholtz P, Allen EE, Ram RJ, Richardson PM et al. (2004). Community structure and metabolism through reconstruction of microbial genomes from the environment. Nature 428: 37–43.
Venter JC, Remington K, Heidelberg JF, Halpern AL, Rusch D, Eisen JA et al. (2004). Environmental genome shotgun sequencing of the Sargasso Sea. Science 304: 66–74.
Wang L, Chen S, Vergin KL, Giovannoni SJ, Chan SW, DeMott MS et al. (2011). DNA phosphorothioation is widespread and quantized in bacterial genomes. Proc Natl Acad Sci USA 108: 2963–2968.
Wang L, Chen S, Xu T, Taghizadeh K, Wishnok JS, Zhou X et al. (2007). Phosphorothioation of DNA in bacteria by dnd genes. Nat Chem Biol 3: 709–710.
Woese CR . (1987). Bacterial evolution. Microbiol Rev 51: 221–271.
Woese CR . (2004). A new biology for a new century. Microbiol Mol Biol Rev 68: 173–186.
Acknowledgements
We thank David Karl and Matt Church, the captain and crew of the R/V Kilo Moana, and Hawaii Ocean Time Series personnel for facilitating work at sea and sample collection. We thank the members of the DeLong lab for many helpful discussions. This work was supported by a grant from the Gordon and Betty Moore Foundation (to EFD), by the Biological and Environmental Research program of the Office of Science in the US Department of Energy (to EFD), and by NSF Science and Technology Center Award EF0424599. This work is a contribution of the Center for Microbial Oceanography: Research and Education (C-MORE).
Author information
Authors and Affiliations
Corresponding author
Additional information
Supplementary Information accompanies the paper on The ISME Journal website
Supplementary information
Rights and permissions
About this article
Cite this article
Danhorn, T., Young, C. & DeLong, E. Comparison of large-insert, small-insert and pyrosequencing libraries for metagenomic analysis. ISME J 6, 2056–2066 (2012). https://doi.org/10.1038/ismej.2012.35
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ismej.2012.35
Keywords
This article is cited by
-
Ecosystem biomonitoring with eDNA: metabarcoding across the tree of life in a tropical marine environment
Scientific Reports (2017)
-
Functional metagenomic screen reveals new and diverse microbial rhodopsins
The ISME Journal (2016)
-
Strong spurious transcription likely contributes to DNA insert bias in typical metagenomic clone libraries
Microbiome (2015)
-
Sequencing platform and library preparation choices impact viral metagenomes
BMC Genomics (2013)