Abstract
The copper membrane monooxygenases (CuMMOs) are an important group of enzymes in environmental science and biotechnology. Areas of relevance include the development of green chemistry for sustainable exploitation of methane (CH4) reserves, remediation of chlorinated hydrocarbon contamination and monitoring human impact in the biogeochemical cycles of CH4 and nitrogen. Challenges for all these applications are that many aspects of the ecology, physiology and structure–function relationships in the CuMMOs are inadequately understood. Here, we describe genetic and physiological characterization of a novel member of the CuMMO family that has an unusual physiological substrate range (C2–C4 alkanes) and a distinctive bacterial host (Mycobacterium). The Mycobacterial CuMMO genes (designated hmoCAB) were amenable to heterologous expression in M. smegmatis—this is the first example of recombinant expression of a complete and highly active CuMMO enzyme. The apparent specific activity of recombinant cells containing hmoCAB ranged from 2 to 3 nmol min–1 per mg protein on ethane, propane and butane as substrates, and the recombinants could also attack ethene, cis-dichloroethene and 1,2-dichloroethane. No detectable activity of recombinants or wild-type strains was seen with methane. The specific inhibitor allylthiourea strongly inhibited growth of wild-type cells on C2–C4 alkanes, and omission of copper from the medium had a similar effect, confirming the physiological role of the CuMMO for growth on alkanes. The hydrocarbon monooxygenase provides a new model for studying this important enzyme family, and the recombinant expression system will enable biochemical and molecular biological experiments (for example, site-directed mutagenesis) that were previously not possible.
Similar content being viewed by others
Introduction
Enzymes in the ammonia monooxygenase superfamily (here termed copper-containing membrane monooxygenases—CuMMOs) are of high biogeochemical and chemical importance (Bedard and Knowles, 1989; Hakemian and Rosenzweig, 2007). Known CuMMOs are restricted to methanotrophs and autotrophic ammonia-oxidizers, nearly all of which grow solely on C1 compounds (Table 1) (Konneke et al., 2005; Stoecker et al., 2006; Dunfield et al., 2007; Baani and Liesack, 2008; Martens-Habbena et al., 2009; Ettwig et al., 2010; Semrau et al., 2010). The resulting ‘restricted role’ paradigm has implications for applications involving CuMMOs. The unusually tight correlation between a single enzyme family and globally important biogeochemical processes such as methane and nitrous oxide flux (Conrad, 1996) suggests CuMMO genes are extremely useful markers of biological feedbacks in global climate change (Singh et al., 2010). However, the distinctive chemistry of CuMMOs and physiological specialization of their native hosts creates major challenges for biochemical characterization of the enzymes (Balasubramanian et al., 2010).
PCR-based environmental surveys have made great progress in identifying the ecological distribution and relevance of CuMMO-containing organisms. Such surveys have repeatedly shown CuMMO sequences are correlated to parameters, such as gas flux, land use and climatic conditions (Menyailo et al., 2008; Avrahami and Bohannan, 2009). However, they have also revealed novel diversity not represented in cultured organisms (Holmes et al., 1999; Leininger et al., 2006; Kip et al., 2010; Luke et al., 2010). These novel environmental sequences pose a challenge for biogeochemical interpretation since our current understanding of structure–function relationships in CuMMOs is too limited to allow us to predict their functions (Singh et al., 2010). This limitation largely reflects the inability to efficiently produce recombinant enzymes or genetically manipulate wild-type strains. These challenges are widely attributed as consequences of both the innate enzyme properties (for example, membrane localization and distinctive metal centers), and the specialized metabolism and genetic intractability of the host bacteria (Semrau et al., 2010). Characterizing the CuMMOs of uncultured organisms is essential to allow functional interpretation of environmental data sets, and also offers the potential to identify more experimentally tractable model organisms.
It has long been postulated that CuMMOs may have roles outside of methanotrophy and ammonia oxidation. The first evidence in this regard came from butane-oxidizing bacteria. Two strains (Nocardioides CF8 and Mycobacterium JOB5) contained a butane monooxygenase activity with similar acetylene and allylthiourea (ATU) inhibition characteristics to the pattern seen for particulate methane monooxygenase (pMMO) and ammonia monooxygenase (AMO) (Hamamura et al., 1999, 2001). In both strains, ATU inhibited butane growth and 14C-labelled acetylene inhibited growth and resulted in labelling of a polypeptide corresponding to the size of a CuMMO subunit. Very recently, genes coding for a three-component monooxygenase with sequence homology to CuMMOs were reported in the draft genome sequence of Nocardioides CF8, and were transcribed in the presence of butane (Sayavedra-Soto et al., 2011). CuMMO sequences have also been linked to ethane-degrading organisms (Genbank accession number: BAH22836). These significant observations suggest that the paradigm of obligate association with physiologically specialized organisms needs revision.
Mycobacterium strain NBB4 was isolated from estuarine sediment by enrichment on ethene as sole carbon source (Coleman et al., 2006). This strain utilizes a broad spectrum of hydrocarbons (C2 to C16 alkanes, C2–C4 alkenes, but not methane) and contains a wide diversity of monooxygenase genes, including four different soluble di-iron monooxygenases (SDIMOs), three cytochrome p-450's and an alkB homolog (Coleman et al., 2011). We previously proved that growth on ethene was linked to one of the SDIMOs (EtnABCD), but we could not identify the monooxygenase (MO) responsible for oxidation of gaseous alkanes. Here, we show that the ‘missing’ gaseous alkane MO of strain NBB4 is a CuMMO, and amenable to recombinant expression in Mycobacterium smegmatis, thus providing a new model system for the study of this important enzyme family.
Materials and methods
Media, bacterial strains and culture conditions
Mycobacterium strains were grown aerobically at 30 °C, using a minimal medium described previously (Coleman et al., 2006), while Escherichia coli (E. coli) strains were grown aerobically at 37 °C in LB medium. Broth media were shaken at 200 r.p.m. Mycobacterium rhodesiae NBB3 and Mycobacterium chubuense NBB4 were isolated in a previous study (Coleman et al., 2006) by enrichment culture on ethene. E. coli EPI300 (F−mcrA Δ(mrr-hsdRMS-mcrBC) Φ80dlacZΔM15 ΔlacX74 recA1 endA1 araD139 Δ(ara, leu)7697 galU galK λ−rpsL (StrR) nupG trfA dhfr) (Epicentre Biotechnologies, Madison, WI, USA), E. coli TOP10 (F−mcrA Δ(mrr-hsdRMS-mcrBC) Φ80lacZΔM15 ΔlacX74 recA1 araD139 Δ(ara leu) 7697 galU galK rpsL (StrR) endA1 nupG) (Invitrogen, Carlsbad, CA, USA) and Mycobacterium smegmatis mc2-155 (Snapper et al., 1990) were used as cloning hosts. Where required for plasmid maintenance, kanamycin (Km) was added at 50 μg ml–1 for E. coli and 20 μg ml–1 for Mycobacterium.
Analytical techniques
Alkanes and chlorinated aliphatic compounds were analyzed by gas chromatography of headspace gas samples (250 μl) using an HP 5890 series II machine (Hewlett-Packard, Wilmington, DE, USA) with a PLOT-Q column, using helium as the carrier gas and flame ionization detection. Quantification was done via an external standard method using a five-point standard curve, with each injection performed in triplicate. Peak areas were converted using the standard curve to amounts expressed as μmoles per bottle. Protein was quantified by an ultraviolet absorption method (Kalb and Bernlohr, 1977) following hot alkaline lysis of the cells (Coleman et al., 2002a). Chloride analysis was done via a colorimetric assay (Coleman et al., 2002b).
Epoxides were detected using a nitrobenzylpyridine assay based on the protocol of Guengerich et al. (1979), as follows. A 2-ml glass vial containing 500 μl of 4-(4-nitrobenzyl)pyridine (100 mM in ethylene glycol) was placed inside a 16 ml bottle containing ethene-grown NBB4 cells (4 ml) at an OD600 of 13±0.5 (0.86 mg protein ml–1). The outer bottle was crimp-sealed with a Teflon-faced butyl rubber stopper, while the inner vial was left open. Substrate (ethene) was added as 10% v/v gas in the headspace, and the bottles incubated at 30 °C with shaking at 200 r.p.m for 24 h. The bottle containing 4-(4-nitrobenzyl)pyridine reagent was removed, and heat-treated (80 °C, 1 h) to enhance conjugate formation, then triethylamine solution (0.5 ml, 1:1 v/v in acetone) was added, and the absorbance at 550 nm was measured immediately. The absorbance was converted to nmol epoxide using the extinction coefficient for the epoxyethane:4-(4-nitrobenzyl)pyridine conjugate (2100 M cm−1) (Morrill et al., 1981).
Effect of inhibitors on growing cultures
The effect of the inhibitors ATU and nitrapyrin was studied by monitoring the change in optical density at 600 nM (OD600) over time in NBB4 cultures growing on C2–C4 alkanes (10% of headspace), ethene (10% of headspace), potassium acetate (20 mM) or glucose (20 mM) in the presence or absence of ATU (25 μM) or nitrapyrin (25 μM), which were added to the medium before inoculation. The effect of copper on growth was studied by comparing growth (as OD600) on the same carbon sources described above in regular minimal medium (0.8 μM Cu2+) or copper-free medium, which was prepared in nitric-acid washed glassware with high purity Milli-Q water (Millipore, Billerica, MA, USA).
Construction, screening and sequencing of fosmid libraries
High molecular weight genomic DNA from strain NBB4 was extracted and purified by a previously described method (Coleman et al., 2002a), then a fosmid library using the pCC1FOS vector was prepared according to the manufacturer's instructions (Epicentre Biotechnologies). Clones were screened for the presence of SDIMOs by PCR as described previously (Coleman et al., 2011), and one clone containing an SDIMO similar to soluble methane monooxygenase was sequenced on both strands by a primer-walking strategy using the Sanger sequencing method at the Australian Genome Research Facility (Westmead node). The sequences were assembled and visualized using the Vector NTI software (Invitrogen). Gene functions were inferred from BLAST searches with the inferred protein sequences against the Genbank and conserved domain databases.
Phylogenetic analysis
Searches of public databases were conducted using BLASTN and BLASTP for hmoCAB and HmoCAB, respectively. In environmental sequence databases and the non-redundant databases, the only hits of >50% identity for any of the three genes or their inferred proteins were recently deposited, unpublished Actinobacterial genome sequences, Nocardioides CF8, and Nocardioides Broad-1. For construction of phylogenetic trees a representative selection of sequences homologous to PmoA, encompassing all major groups identified in four recent studies was used (Ettwig et al., 2010; Kip et al., 2010; Luke et al., 2010; Tavormina et al., 2010). An ungapped alignment corresponding to the region between Lys59 and Thr207 of Mc. capsulatus Bath PmoA was used for analyses.
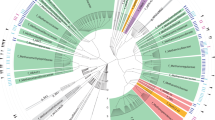
Both protein and nucleotide sequences were used for construction of trees using programs available in the Phylip and Geneious packages (http://www.geneious.com). Many CuMMO-containing organisms contain multiple non-orthologous alleles, so this dendrogram does not reflect the phylogenetic relationships of the parent bacterial strains. The isolated phylogenetic position of the Hmo proteins was seen with all methods of analysis, and for all Hmo subunits (trees derived from HmoB and HmoC analysis were not shown here because no sequence data are available for these subunits in most environmental sequence groups). The tree shown in Figure 2 is a distance matrix tree of HmoA homologs constructed using Protdist (JTT matrix) and Fitch, with the Nitrosopumilis mirabilis AmoA sequence (ABY89319) used as an outgroup. Note that this outgroup sequence cannot be unambiguously aligned over the full length of the alignment and accordingly the position of this sequence is shown as a dimensionless line. Note that the Proline-169 residue of the M. oxyfera sequence (CBE69519) has no homologous position in all other bacterial sequences—this site was deleted from the alignment.
Protein accession numbers for sequences included in the tree shown in Figure 2 are: M84-P22 group (1_ABC86668, 2_ACD68272, 3_ACD68273, 4_ACM46919, 5_ACD68292, 6_CAI30606); Candidatus Methylomirabilis oxyfera (7_CBE69519); Verrucomicrobia PmoA1/PmoA2 group (8_ACK55191, 9_ABX56601, 10_ACK55192, 11_ABX56604); alphaProteobacteria methanotrophs group (12_AAD47927, 13_AAV67717, 14_AAL86710, 15_CAC45630, 16_CAD24093, 17_CAI30620, 18_CAD30373, 19_CAD30371, 20_CAD30381, 21_CAD24094, 22_CAD24092); gammaProteobacteria methanotroph PmoA group (23_ACS72359, 24_CAE22498, 25_AAG13081, 26_AAC61804, 27_AAX48776, 28_AAG13080, 29_AAL09399, 30_AAF08215, 31_AAA87218, 32_AAX37294, 33_AAB49821, 34_ADM12710, 35_AAC04380); gammaProteobacteria AOB group (36_AAV67698, 37_AAV67707, 38_AAV67689, 39_AAC25091); TUSC group (40_AAZ06193, 41_AAZ06151, 42_CAC69466); gammaProteobacteria ET-HIRO (43_BAH22836, 44_ACC85973); RA21 group (45_CAC84847, 46_ACJ54056, 47_AAL87425, 48_AAD47928); gp23 clone 49_AAK51669; MR1 group (50_ACS94422, 51_ACM46913, 52_AAF19026); gammaProteobacteria methanotroph PxmA group (53_ABL85665, 54_ACE95888, 55_ACJ53965, 56_ACE95881, 57_ACE95890); M84-P105 group (58_CBH32423, 59_CAC69467, 60_CAC69468, 61_ACJ53974, 62_ACJ54003, 63_CAZ15776, 64_AAF64113); betaProteobacteria AOB group (65_AAA66194, 66_CAA62336, 67_AAB38710, 68_AAC25057); P13.9 group (69_ABU45542, 70_AAL87436, 71_ACM46931, 72_ACM46953); Candidatus Crenothrix PmoA (73_ABC59825; 74_ABC59822; 75_ABC59827); Verrucomicrobia PmoA3 group (76_ACK55193, 77_ABX56597); Actinobacterial group (NBB4_ADT71671; NBB3_draft genome; CF8_ADW80613; Broad_EGD43187).
Cloning of hmoCAB genes into pMycoFos to make pHMO
A newly developed Mycobacterium–E. coli shuttle fosmid vector (pMycoFos, 12.5 kb, KmR) was used to clone and express the hmoCAB genes (Figure 5). Full details of the construction and evaluation of this vector are reported elsewhere (Ly and Coleman, 2011), but in brief, this vector combines the E. coli replication and stabilization functions of the pCC1FOS fosmid (Epicentre Biotechnologies) (Wild et al., 2002) with the Mycobacterium replicon and acetamidase promoter regions from the pJAM2 shuttle vector (Triccas et al., 1998)—this allows control of the copy number in E. coli via arabinose induction, and expression of cloned genes in Mycobacterium via acetamide induction.
The hmoCAB genes of NBB4 were amplified from genomic DNA using primers NVC307 (5′-TTTATTTAAATCTGCCAGCTAACAATCTCG-3′) and NVC308 (5′-TTTATTTAAATCTATTGGGTTCCGTATTCAGG-3′) containing SwaI sites (underlined). The resulting PCR product (3.2 kb) was digested with SwaI, ligated with similarly digested pMycoFos vector, and transformed into chemically competent E. coli EPI300 cells (Epicentre Biotechnologies) by heat shock. After recovery and plating on LB–Km agar, positive clones containing hmoCAB were detected by PCR using primers HEO3 (5′-CAGCCAGGATAAAAGTCTCGGGC-3′) (binds in hmoA) and NW211 (5′-GCTCATAACACCCCTTGTATTACTG-3′) (binds in KmR gene of vector), which yields a 2.2 kb product if the insert DNA had ligated correctly into pMycoFos. One positive clone (designated pHMO) was retained for further work, and its structure confirmed by sequencing the ligation junctions, and by restriction digests using SwaI, EcoRI, BamHI, HindIII and KpnI. The pHMO plasmid was transformed into electrocompetent cells of strain mc2-155 by electroporation (Bio-Rad GenePulser, Hercules, CA, USA; 800 ohms, 2.5 μF, 2.5 kV). After recovery in 1 ml LB broth for 4 h at 30 °C, transformants were plated on LB–Km agar and incubated at 30 °C for 7 days. All subsequent activity assays were done with freshly transformed cells of mc2-155(pHMO) (that is, not from glycerol stocks).
Activity assays with resting cell suspensions
The activity of the cloned hmoCAB genes was quantified in resting cell suspensions using gas chromatography–flame ionization detection (alkanes, chlorinated aliphatics), colorimetric analysis of epoxides (ethene) and/or chloride analysis (chlorinated aliphatics). Cells of mc2-155(pHMO) were grown in 30 ml minimal medium supplemented with glucose (20 mM), Tween-80 (0.05 % v/v), acetamide (0.2 % v/v) and Km (20 μg ml–1) to an OD600 of 1.0. The cells were washed twice in potassium phosphate buffer (20 mM K2HPO4, pH 7.0) containing Tween-80 then resuspended in 4 ml potassium phosphate buffer containing 20 mM glucose and Tween-80, and transferred to a gas-tight 16 ml serum bottle, and crimp-sealed with a Teflon-faced butyl rubber stopper, and aluminum crimp seal cap. Glucose was added to the cell suspensions as preliminary tests (not shown) indicated that higher activity could be maintained in the resting cells when they were provided with a supplemental energy source, but it should be noted that cells could not grow under the conditions of this assay because they lacked a nitrogen source and trace metals.
Test substrate (2 μmol methane, ethane, ethene, propane, butane, cis-dichloroethene or 1,2-dichloroethane) was added either as neat gas (alkanes) or from a 20 mM aqueous stock solution (chlorinated compounds) and the suspensions were incubated at 30 °C with shaking at 200 r.p.m. In the case of alkanes and chlorinated aliphatics, headspace samples (250 μl) were taken at intervals for gas chromatography–flame ionization detection analysis. For alkanes, degradation rates were calculated from the initial linear part of the substrate depletion curves, and then the rates were converted to apparent specific activities, expressed as nmol substrate consumed min–1 per mg protein. For chlorinated aliphatics, rates were not measured, but instead, an end-point assay was used where the amount of substrate consumed and the amount of chloride released were calculated after overnight incubation (16 h). In the case of ethene, many identical bottles were set up and one was destroyed at each time point for 4-(4-nitrobenzyl)pyridine assay, as described above, and the activity was expressed as nmol epoxide formed min–1 per mg protein.
All activity assays included controls consisting of mc2-155 cells carrying the vector only, with no CuMMO gene insert. All experiments were repeated at least three times, and the average apparent specific activity was calculated.
Results and Discussion
Detection of copper monooxygenase genes in Mycobacterium NBB4
Our initial interest in strain NBB4 concerned its unusual complement of SDIMOs. A fosmid library of NBB4 genomic DNA was screened for clones that contained monooxygenase genes as previously described (Coleman et al., 2011). Additional sequencing of one SDIMO-containing fosmid led to the identification of a CuMMO gene cluster with homology to pMMO and AMO (each predicted protein 30–40% identity to known bacterial pMMO or AMO proteins). The full sequence of this fosmid insert (35 662 bp) has been deposited in GenBank with accession number GU174751. The CuMMO genes are henceforth referred to as hmoCAB (hydrocarbon monooxygenase); the gene names chosen correspond to the subunits of pMMO and AMO, because the arrangement of the hmoCAB subunits was identical to that of amoCAB and pmoCAB. A schematic diagram of the hmoCAB genomic region of strain NBB4 and a similar gene cluster observed in the draft genome of the closely related alkene/alkane-degrading Mycobacterium rhodesiae NBB3 are shown in Figure 1. A detailed annotation of the strain NBB4 region is shown in Supplementary Table 1. Surprisingly, despite the very close phylogenetic relationship of the two Mycobacterium strains (NBB3 and NBB4) the level of homology between their CuMMO genes is only moderate (61%, 68%, 52% amino acid identity in HmoC, HmoA, HmoB, respectively).
Schematic diagram of the genomic context of hmoCAB genes in strains NBB4 and NBB3. Details of NBB4 gene annotations are in Supplementary Table S1. Regions of synteny with strain NBB3 are indicated by shaded blocks. The two indicated NBB3 predicted genes show homology to NBB4 genes outside the displayed region (Supplementary data).
Two lines of evidence led us to postulate that hmoCAB and related enzymes are involved in alkane metabolism. First, biochemical evidence for the presence of CuMMOs in Actinobacteria with a physiological role in butane degradation had previously been reported in Nocardioides CF8 and Mycobacterium JOB5 (Hamamura et al., 1999, 2001). Very recently, the presence of CuMMO genes in strain CF8 was confirmed in the draft genome sequence (Sayavedra-Soto et al., 2011). We also note that CuMMO homologs are present in the draft sequence of a presumed Nocardioides organism (Broad-1) serendipitously obtained during sequencing of the fungus Coccidioides (gb:EGD43186). Second, in our NBB4 fosmid sequence, genes encoding alcohol dehydrogenase and acyl-CoA ligase homologs were found in close proximity to hmoCAB—these potentially perform the second and third steps in an alkane assimilation pathway. A range of bioinformatic and physiological experiments were conducted to test the hypothesis that, with respect to previously known CuMMOs, the Actinobacterial enzymes comprise a new subgroup with a distinct physiological role.
The HmoCAB enzymes of NBB4 and NBB3 were shown to have a deep-branching position within the bacterial radiation of CuMMOs (Figure 2). Together with the CuMMO homologs of strain CF8 and Broad-1 they represent a group with a distinct evolutionary history that we refer to as the Actinobacterial copper monooxygenases. Comparative sequence analysis suggests these enzymes all share the key structural attributes of the CuMMOs. Structures of the pMMO of Ms. trichosporium and Mc. capsulatus have identified two metal centers whose ligands are strongly conserved in other CuMMOs (Figure 3). A metal center in PmoB has been shown to be a dinuclear copper site coordinated by three His residues and has been implicated in methane oxidation (Balasubramanian et al., 2010). All the ligands of this copper site are conserved in HmoCAB, and the surrounding region also shows other conserved residues (Figure 3). The nature and role of the metal center in PmoA/PmoC is more controversial. The three residues identified as metal ligands in the pMMO structures of Mc. capsulatus (Lieberman and Rosenzweig 2005) and Ms. trichosporium (Hakemian et al., 2008) are all conserved in HmoCAB. However, the nature of the metal varies (zinc or copper), and it has also been postulated that in pMMOs this site could be a di-iron site with nearby carboxylate residues also involved in metal co-ordination (Semrau et al., 2010). An interesting feature of the predicted HmoCAB proteins is that an aspartate residue postulated to be important in the PmoA/PmoC site is not conserved in HmoCAB, a trait it shares with the AMOs.
Relationship of the NBB4 HmoCAB enzyme to other Bacterial CuMMOs based on PmoA homologs. Tree was constructed from an ungapped alignment of 149 residues. Numbers at each tip refer to the sequence used for tree construction. Accession numbers and details of tree construction are given in the Materials and methods section. Environmental sequences are shown in grey. Branches leading to a sequence derived from an organism of known physiology and phylogenetic affiliation are shown in bold. These include: Alkane-degrading heterotrophs, Proteobacterial methanotrophs, Proteobacterial ammonia oxidizers, Verrucomicrobial methanotrophs and the candidate division NC10 anaerobic methanotroph. The disjunct distribution of sequences from the same phylogenetic groups highlights that CuMMO sequences are not all orthologous.
Conservation of the metal ligand-binding sites of pMMO in HmoCAB and other CuMMOs. Accession numbers for PmoB and PmoA proteins are given where available. Isologous sites to ligands of the metal centers identified in the published structures of pMMO are indicated as follows: The PmoB dinuclear copper site is shown with an asterisk (*); the PmoA/PmoC variable metal site with a hash (#) and; the carboxylate side-chain residues postulated to co-ordinate a di-iron center at the variable metal site are marked with a plus (+). Note: the Np. maritimus PmoA sequence is shown in lower case because the homology of this protein is not established.
Growth experiments indicate a CuMMO is required for growth on small alkanes
Available sequence data indicate the strain NBB4 genome has at least nine distinct monooxygenases, all of which could conceivably encompass small alkanes within their substrate range (Coleman et al., 2011). Several lines of evidence argue that the eight iron-containing enzymes are not involved in initiating growth on gaseous hydrocarbons. A copper requirement is a distinctive feature of the CuMMO-dependent autotrophic ammonia oxidizers and methanotrophs (Nielsen et al., 1997; Lieberman and Rosenzweig, 2005), therefore we tested the effect of copper-free media vs standard media (0.8 μM Cu2+) on the growth substrate range of strain NBB4. In copper-free media, growth on gaseous alkanes (ethane, propane and butane) was strongly limited, but growth on other carbon sources, including alkenes, organic acids and glucose, was unaffected (Figure 4). This indicates a specific physiological requirement for copper for growth on alkanes as sole carbon source, and that this is likely for enzymes involved in the initial degradative steps.
Copper requirement and ATU sensitivity of Mycobacterium strain NBB4 for growth on different substrates. (a) Final growth yield (96 h) in minimal medium with 0.8 μM Cu2+ or in the absence of added copper. (b) Final growth yield (96 h) in minimal medium in the presence and absence of 25 μM ATU. For both (a) and (b), data are the average of three replicate experiments, and error bars indicate 1 s.d.
ATU is a mechanistic inhibitor of CuMMO-dependent growth of methanotrophs and autotrophic ammonia oxidizers (Bedard and Knowles 1989; McCarty, 1999). When ATU was added to normal (copper-containing) media, strong growth inhibition was seen for ethane, propane and butane, but not for growth on acetate or ethene (Figure 1). This indicates it is unlikely any of the known alternate MOs in strain NBB4 are essential for initiating growth on alkanes, since there is no precedent for ATU inhibition of an iron-containing MO, or a copper requirement for iron MO-mediated growth on hydrocarbons. Experimental data showing a lack of ATU sensitivity exist for the sMMO of Methylosinus trichosporium and the butane monooxygenase of Thauera butanivorans (Nielsen et al., 1997; Hamamura et al., 2001), and in strain NBB4, SDIMO-mediated growth on ethene was unaffected by ATU. Further indirect support for lack of involvement of iron-MOs comes from the apparent absence of any iron MO from the proteome of NBB4 cells grown on C2–C4 alkanes (Coleman et al., 2011).
Heterologous expression proves HmoCAB is an alkane monooxygenase
We used a heterologous expression approach to link the hmoCAB genes to metabolism of particular substrates. CuMMO genes are notoriously difficult to clone in heterologous hosts (Semrau et al., 1995; Balasubramanian et al., 2010) and our initial attempts using standard E. coli vectors or E. coli–Mycobacterium shuttle vectors were unsuccessful (data not shown). To overcome these difficulties, we developed a novel fosmid-based shuttle vector, with copy number and expression control in E. coli and acetamide-inducible expression of cloned genes in Mycobacterium—this enabled the stable cloning of the hmoCAB operon of strain NBB4 yielding the plasmid pHMO (Figure 5). Initial characterization of pHMO (sequencing of the junctions between insert and vector) confirmed that the orientation and boundaries of the cloned PCR product were correct. This construct was used to perform the physiological tests described below. Subsequent analysis of the plasmid structure confirmed that no errors had been introduced into the sequence of hmoCAB during the pCR-mediated cloning but also indicated that two complete copies of the hmoCAB gene clusters had been cloned, with the boundary being the SwaI site of the PCR primers. We have subsequently generated a single copy version and confirmed its activity (data not shown).
Structure of pHMO construct. The backbone of the construct is the pMycoFos vector, full details of which are described elsewhere (Ly and Coleman, 2011). Two copies of the hmoCAB gene cluster (3 187 bp) were cloned into pMycoFos to create pHMO (18 908 bp). The structure of pHMO has been validated by sequencing of the insert-vector junctions and by restriction mapping.
No alkane monooxygenase activity was seen in cells of E. coli EPI300(pHMO), but transfer of pHMO into Mycobacterium smegmatis mc2-155 yielded activity. Gas chromatography assays of substrate depletion (Figure 6) indicated that resting cell suspensions had apparent specific activities of 2.1, 2.9 and 2.2 nmol min–1 per mg protein for ethane, propane and butane, respectively. These activities were 10–20-fold lower than those of wild-type NBB4 cells grown on the short chain alkanes as sole carbon source (Coleman et al., 2011). No activity of mc2-155(pHMO) cells was observed toward methane, and no activity was seen in mc2-155(pMycoFos) cells lacking hmoCAB. This latter control is important because the M. smegmatis mc2-155 genome encodes a putative endogenous aromatic/alkane monooxygenase (Furuya et al., 2011), however, we have not seen evidence of this activity, possibly due to a point mutation in our lineage of strain mc2-155 (unpublished data). There are only two previous reports of successful heterologous expression of CuMMO proteins, but in one case (pMMO expression in Rhodococcus erythropolis) the level of activity was very low (∼0.2 nmol min–1 per mg protein) (Gou et al., 2006), and in the other case (expression of the N-terminal domain of PmoB in E. coli), only a soluble fragment was expressed (Balasubramanian et al., 2010).
Activity of recombinant HMO toward alkanes, alkenes and chlorinated compounds. Panels (a–d) show substrate depletion for gaseous alkanes over time by resting cells of M. smegmatis mc2-155 carrying either pHMO (triangles) or the vector pMycoFos with no insert (squares). For all substrates, data are the average of three independent experiments, and error bars indicate 1 s.d. The data points between 0–180 min were used in each case for calculation of specific activities (linear regressions not shown). Panel (e) shows production of epoxyethane from ethene. Panel (f) shows metabolism of cis-dichloroethene (cDCE) and 1,2-dichloroethane (DCA). Data shown are amount of substrate depleted (white) and amount of chloride released (dark) measured at completion of the timecourse.
HmoCAB also oxidizes ethene to epoxyethane and dechlorinates pollutants
Cells of mc2-155(pHMO) also displayed activity with ethene and chlorinated aliphatic compounds. Production of epoxyethane from ethene was detected using a colorimetric assay (Figure 6), and an activity of 3.2 nmol min–1 per mg protein was calculated based on the rate of epoxide formation, which agrees well with the rates of alkane oxidation observed above. There is an interesting parallel here with the dual-MMO methanotrophs. We have previously demonstrated that growth of NBB4 on ethene is associated with the SDIMO enzyme EtnABCD, as is the case in other ethenotrophic bacteria (Mattes et al., 2010; Coleman et al., 2011). The observation that HmoCAB is also capable of ethene oxidation indicates that this bacterium has distinct enzymes potentially capable of initiating growth on ethene. In dual MMO methanotrophs, cells switch between growth using an SDIMO (sMMO) and a CuMMO (pMMO) according to the availability of copper. There is no obvious effect of copper on cell growth rate on ethene as sole carbon source, but this possibility requires further investigation (for example, to test if etnABCD knockout mutants can still grow on ethene).
Wild-type cells of strain NBB4 attack a wide range of pollutant compounds (Le and Coleman, 2011). Recombinant HMO-expressing cells of mc2-155 also attacked the chlorinated aliphatic compounds cis-dichloroethene and 1,2-dichloroethane (Figure 6), which are common groundwater pollutants (Squillace et al., 1999). We did not calculate specific activities with the chlorinated compounds (an end point assay was used), but we did measure inorganic chloride in these experiments, which revealed chloride release from both cis-dichloroethene and 1,2-dichloroethane, confirming that carbon–chlorine bonds in these molecules were cleaved. The stoichiometry of chloride release indicated that only 0.3–0.5 mol chloride per mol substrate was released, and thus that these substrates were not mineralized. It is notable that in other work with ethane-grown wild-type cells of strain NBB4 (Le and Coleman, 2011), the stoichiometry of chloride release from cis-dichloroethene and 1,2-dichloroethane was different—the reasons for this are unclear, but are likely to include the involvement of additional enzymes from the MO repertoire of strain NBB4. It is evident that part of the chlorinated hydrocarbon degradative capacity of alkane-grown NBB4 cells can be attributed to HMO.
Conclusions
Multiple lines of evidence (comparative sequence analysis, ATU inhibition, copper requirement and recombinant expression) indicate that the HmoCAB enzyme of strain NBB4 is a CuMMO, with C2–C4 alkanes as the physiological substrates. Phylogenetic analyses, along with available physiological data, suggest that HmoCAB of NBB4 is a member of a functionally similar group of enzymes that are present in disparate heterotrophic Actinobacteria, where they enable opportunistic utilization of alkanes. Another member of this group is also likely to be present in the propane oxidizer Mycobacterium JOB5 (Hamamura et al., 1997, 1999; Sayavedra-Soto et al., 2011). Two aspects of genome relationships in this group are especially significant; the sequence divergence between these Actinobacterial CuMMOs is large relative to the ribosomal RNA divergence of the parent organisms, and available genome sequence data indicate the genes are not universally present among closely related bacteria. These are in strong contrast to the Proteobacterial methanotrophs and AOB where the genes encoding pMMO and AMO fit the concept of extended core genes (Lapierre and Gogarten, 2009). That is, they occur in the genomes of nearly all members of an evolutionary group (four separate groups within the Proteobacteria in this case) and are under high selective pressure (slow rates of divergence). The Actinobacterial HMO group are non-essential genes, that fit the concept of character genes (or accessory genes in other definitions). Such genes are variably present within a lineage, evolve more rapidly and are far more prone to horizontal gene transfer. Our data, therefore, demonstrate that the CuMMO paradigm needs to be modified to reflect that members of this family are not always core genes in physiologically specialized organisms.
The Actinobacterial group CuMMOs are almost certainly relevant to short chain hydrocarbon degradation and bioremediation and potentially also significant for heterotrophic nitrification (Hamamura et al., 1999). Genes for the Actinobacterial CuMMO group are not successfully amplified using extant pMMO or AMO primers (Holmes et al., 1995; Rotthauwe et al., 1997; Santoro et al., 2010). This situation, where an entire functional group has been overlooked, is reminiscent of the discovery of the ammonia-oxidizing archaea (Konneke et al., 2005) and subsequent demonstration that archaea (not bacteria) are the dominant nitrifiers in many environments (Leininger et al., 2006; Martens-Habbena et al., 2009; Zhang et al. 2010). A further consideration for biogeochemical studies is that ecological extrapolations from CuMMO sequences depend on congruence between CuMMO sequence relationships, ribosomal RNA phylogeny and organism physiology (Singh et al., 2010). This congruence is strong for the characterized methanotrophs and ammonia-oxidizers where CuMMOs are core genes, but unlikely to hold true for C2–C4 alkane-degraders, because of the diverse MO types that can act on C2–C4 alkanes, and the fact that distribution of the CuMMOs in the Actinobacteria lineage appears to be sporadic.
The recombinant expression system we have developed for the HmoCAB holoenzyme opens up new approaches for CuMMO biochemistry (for example, site-directed mutagenesis), and we anticipate it will help resolve questions regarding the catalytic roles of the different protein subunits. Of particular relevance here is that the nature of the metal-containing sites in CuMMOs remains controversial. The PmoA/AmoA subunit was long thought to be the site of catalysis since acetylene, a suicide substrate, binds to this protein (Zahn and DiSpirito, 1996; Gilch et al., 2009). However, when structures of pMMO were solved, two metal sites were revealed (Lieberman and Rosenzweig, 2005; Hakemian and Rosenzweig, 2007) and recently the PmoB site was confirmed as having methane hydroxylase activity when the PmoB subunit of the M. capsulatus pMMO was expressed in soluble form in E. coli (Balasubramanian et al., 2010). However, a paradox remains since the ligands for the PmoB site are not conserved across all methanotrophs (Dunfield et al., 2007; Pol et al., 2007), and this experiment does not explain the canonical membrane association of CuMMOs. The availability of a recombinant expression system for the holoenzyme creates new opportunities to probe the nature of the metal centers in this enzyme family. We anticipate it will also be useful for characterizing different CuMMO alleles in organisms containing heterologous copies (Dunfield et al., 2007; Pol et al., 2007; Baani and Liesack, 2008), and for determining the function of environmental CuMMO gene clusters (Dumont et al., 2006).
References
Avrahami S, Bohannan BJM . (2009). N2O emission rates in a California meadow soil are influenced by fertilizer level, soil moisture and the community structure of ammonia-oxidizing bacteria. Glob Change Biol 15: 643–655.
Baani M, Liesack W . (2008). Two isozymes of particulate methane monooxygenase with different methane oxidation kinetics are found in Methylocystis sp strain SCZ. Proc Natl Acad Sci USA 105: 10203–10208.
Balasubramanian R, Smith SM, Rawat S, Yatsunyk LA, Stemmler TL, Rosenzweig AC . (2010). Oxidation of methane by a biological dicopper centre. Nature 465: 115–121.
Bedard C, Knowles R . (1989). Physiology, biochemistry, and specific inhibitors of Ch4, Nh4+, and co-oxidation by methanotrophs and nitrifiers. Microbiol Revs 53: 68–84.
Coleman NV, Mattes TE, Gossett JM, Spain JC . (2002a). Phylogenetic and kinetic diversity of aerobic vinyl chloride-assimilating bacteria from contaminated sites. Appl Environ Microbiol 68: 6162–6171.
Coleman NV, Mattes TE, Gossett JM, Spain JC . (2002b). Biodegradation of cis-dichloroethene as the sole carbon source by a beta-proteobacterium. Appl Environ Microbiol 68: 2726–2730.
Coleman NV, Bui NB, Holmes AJ . (2006). Soluble di-iron monooxygenase gene diversity in soils, sediments and ethene enrichments. Environ Microbiol 8: 1228–1239.
Coleman NV, Yau S, Wilson NL, Nolan LM, Migocki MD, Ly M et al. (2011). Untangling the multiple monooxygenases of Mycobacterium chubuense strain NBB4, a versatile hydrocarbon degrader. Environ Microbiol Reports 3: 297–307.
Conrad R . (1996). Soil microorganisms as controllers of atmospheric trace gases (H-2, CO, CH4, OCS, N2O, and NO). Microbiol Revs 60: 609–640.
Dumont MG, Radajewski SM, Miguez CB, McDonald IR, Murrell JC . (2006). Identification of a complete methane monooxygenase operon from soil by combining stable isotope probing and metagenomic analysis. Environ Microbiol 8: 1240–1250.
Dunfield PF, Yuryev A, Senin P, Smirnova AV, Stott MB, Hou SB et al. (2007). Methane oxidation by an extremely acidophilic bacterium of the phylum Verrucomicrobia. Nature 450: 879–882.
Ettwig KF, Butler MK, Le Paslier D, Pelletier D, Mangenot S, Kuypers MMM et al. (2010). Nitrite-driven anaerobic methane oxidation by oxygenic bacteria. Nature 464: 543–548.
Furuya T, Hirose S, Osanai H, Semba H, Kino K . (2011). Identification of the monooxygenase gene clusters responsible for the regioselective oxidation of phenol to hydroquinone in mycobacteria. Appl Environ Microbiol 77: 1214–1220.
Gilch S, Vogel M, Lorenz MW, Meyer O, Schmidt I . (2009). Interaction of the mechanism-based inactivator acetylene with ammonia monooxygenase of Nitrosomonas europaea. Microbiology 155: 279–284.
Gou ZX, Xing XH, Luo MF, Jiang H, Han B, Wu H et al. (2006). Functional expression of the particulate methane mono-oxygenase gene in recombinant Rhodococcus erythropolis. FEMS Microbiol Letts 263: 136–141.
Guengerich FP, Crawford WM, Watanabe PG . (1979). Activation of vinyl-chloride to covalently bound metabolites—roles of 2-chloroethylene oxide and 2-chloroacetaldehyde. Biochemistry 18: 5177–5182.
Hakemian AS, Rosenzweig AC . (2007). The biochemistry of methane oxidation. Ann Rev Biochem 76: 223–241.
Hakemian AS, Kondapalli KC, Telser J, Hoffman BM, Stemmler TL, Rosenzweig AC . (2008). The metal clusters of particulate methane monooxygenase from Methylosinus trichosporium OB3b. Biochem 47: 6793–6801.
Hamamura N, Page C, Long T, Semprini L, Arp DJ . (1997). Chloroform cometabolism by butane-grown CF8, Pseudomonas butanovora, and Mycobacterium vaccae JOB5 and methane-grown Methylosinus trichosporium OB3b. Appl Environ Microbiol 63: 3607–3613.
Hamamura N, Storfa RT, Semprini L, Arp D . (1999). Diversity in butane monooxygenases among butane-grown bacteria. Appl Environ Microbiol 65: 4586–4593.
Hamamura N, Yeager CM, Arp DJ . (2001). Two distinct monooxygenases for alkane oxidation in Nocardioides sp strain CF8. Appl Environ Microbiol 67: 4992–4998.
Holmes AJ, Costello A, Lidstrom ME, Murrell JC . (1995). Evidence that particulate methane monooxygenase and ammonia monooxygenase may be evolutionarily related. FEMS Microbiol Letts 132: 203–208.
Holmes AJ, Roslev P, McDonald IR, Iversen N, Henriksen K, Murrell JC . (1999). Characterization of methanotrophic bacterial populations in soils showing atmospheric methane uptake. Appl Environ Microbiol 65: 3312–3318.
Kalb VF, Bernlohr RW . (1977). A new spectrophotometric assay for protein in cell extracts. Anal Biochem 82: 362–371.
Kip N, van Winden JF, Pan Y, Bodrossy L, Reichart GJ, Smolders AJP et al. (2010). Global prevalence of methane oxidation by symbiotic bacteria in peat-moss ecosystems. Nat Geoscience 3: 617–621.
Konneke M, Bernhard AE, de la Torre JR, Walker CB, Waterbury JB, Stahl DA . (2005). Isolation of an autotrophic ammonia-oxidizing marine archaeon. Nature 437: 543–546.
Lapierre P, Gogarten JP . (2009). Estimating the size of the bacterial pan-genome. Trends Genet 25: 107–110.
Le N, Coleman NV . (2011). Biodegradation of vinyl chloride, cis-dichloroethene and 1,2-dichloroethane in the alkene/alkane-oxidising Mycobacterium strain NBB4. Biodegradation. doi:10.1007/s10532-1011-9466-0.
Leininger S, Urich T, Schloter M, Schwark L, Qi J, Nicol GW et al. (2006). Archaea predominate among ammonia-oxidizing prokaryotes in soils. Nature 442: 806–809.
Lieberman RL, Rosenzweig AC . (2005). Crystal structure of a membrane-bound metalloenzyme that catalyses the biological oxidation of methane. Nature 434: 177–182.
Luke C, Krause S, Cavigiolo S et al. (2010). Biogeography of wetland rice methanotrophs. Environ Microbiol 12: 862–872.
Ly MA, Coleman NV . (2011). Construction and evaluation of pMycoFos, a fosmid shuttle vector for Mycobacterium spp. with inducible gene expression and copy number control. J Microbiol Meth. doi:10.1016/j.mimet.2011.06.005.
Martens-Habbena W, Berube PM, Urakawa H, de la Torre JR, Stahl DA . (2009). Ammonia oxidation kinetics determine niche separation of nitrifying Archaea and Bacteria. Nature 461: 976–979.
Mattes TE, Alexander AK, Coleman NV . (2010). Aerobic biodegradation of the chloroethenes: pathways, enzymes, ecology, and evolution. FEMS Microbiol Revs 34: 445–475.
McCarty GW . (1999). Modes of action of nitrification inhibitors. Biol Fert Soils 29: 1–9.
Menyailo OV, Hungate BA, Abraham WR, Conrad R . (2008). Changing land use reduces soil CH4 uptake by altering biomass and activity but not composition of high-affinity methanotrophs. Glob Change Biol 14: 2405–2419.
Morrill TC, Friedrich LE, Machonkin MA, Whitbourne JE, Eastman CA . (1981). N-(2-hydroxyethyl)-4-(p-nitrobenzylidene)-1,4-dihydropyridine (DHP) from the reaction of 4-(p-nitrobenzyl)pyridine (NBP) with ethylene oxide. J Heterocyclic Chem 18: 1645–1647.
Nielsen AK, Gerdes K, Murrell JC . (1997). Copper-dependent reciprocal transcriptional regulation of methane monooxygenase genes in Methylococcus capsulatus and Methylosinus trichosporium. Mol Microbiol 25: 399–409.
Pol A, Heijmans K, Harhangi HR, Tedesco D, Jetten MSM, Op den Camp HJM . (2007). Methanotrophy below pH1 by a new Verrucomicrobia species. Nature 450: 874–878.
Rotthauwe JH, Witzel KP, Liesack W . (1997). The ammonia monooxygenase structural gene amoA as a functional marker: Molecular fine-scale analysis of natural ammonia-oxidizing populations. Appl Environ Microbiol 63: 4704–4712.
Santoro AE, Casciotti KL, Francis CA . (2010). Activity, abundance and diversity of nitrifying archaea and bacteria in the central California Current. Environ Microbiol 12: 1989–2006.
Sayavedra-Soto LA, Hamamura N, Liu C-W, Kimbrel JA, Chang JH, Arp DJ . (2011). The membrane-associated monooxygenase in the butane-oxidizing Gram-positive bacterium Nocardioides sp. strain CF8 is a novel member of the AMO/PMO family. Environ Microbiol Reports. doi:10.1111/j.1758-2229.2010.00239.x.
Semrau JD, Chistoserdov A, Lebron J, Costello A, Davagnino J, Kenna E et al. (1995). Particulate methane monooxygenase genes in methanotrophs. J Bacteriol 177: 3071–3079.
Semrau JD, DiSpirito AA, Yoon S . (2010). Methanotrophs and copper. FEMS Microbiol Revs 34: 496–531.
Singh BK, Bardgett RD, Smith P, Reay DS . (2010). Microorganisms and climate change: terrestrial feedbacks and mitigation options. Nat Revs Microbiol 8: 779–790.
Snapper SB, Melton RE, Mustafa S, Kieser T, Jacobs Jr WR . (1990). Isolation and characterization of efficient plasmid transformation mutants of Mycobacterium smegmatis. Mol Microbiol 4: 1911–1919.
Squillace PJ, Moran MJ, Lapham WW, Price CV, Clawges RM, Zogorski JS . (1999). Volatile organic compounds in untreated ambient groundwater of the United States, 1985–1995. Environ Sci Technol 33: 4176–4187.
Stoecker K, Bendinger B, Schoning B, Nielsen PH, Nielsen JL, Baranyi C et al. (2006). Cohn′s Crenothrix is a filamentous methane oxidizer with an unusual methane monooxygenase. Proc Natl Acad Sci USA 103: 2363–2367.
Tavormina PL, Ussler W, Joye SB, Harrison BK, Orphan VJ . (2010). Distributions of putative aerobic methanotrophs in diverse pelagic marine environments. ISME J 4: 700–710.
Triccas JA, Parish T, Britton WJ, Gicquel B . (1998). An inducible expression system permitting the efficient purification of a recombinant antigen from Mycobacterium smegmatis. FEMS Microbiol Lett 167: 151–156.
Wild J, Hradecna Z, Szybalski W . (2002). Conditionally amplifiable BACs: switching from single-copy to high-copy vectors and genomic clones. Genome Res 12: 1434–1444.
Zahn JA, DiSpirito AA . (1996). Membrane-associated methane monooxygenase from Methylococcus capsulatus (Bath). J Bacteriol 178: 1018–1029.
Zhang LM, Offre PR, He JZ, Verhamme DT, Nicol GW, Prosser JI . (2010). Autotrophic ammonia oxidation by soil Thaumarchaea. Proc Natl Acad Sci USA 107: 17240–17245.
Acknowledgements
This work was supported by grants from the Australian Research Council to NVC and AJH.
Author information
Authors and Affiliations
Corresponding author
Additional information
Supplementary Information accompanies the paper on The ISME Journal website
Rights and permissions
About this article
Cite this article
Coleman, N., Le, N., Ly, M. et al. Hydrocarbon monooxygenase in Mycobacterium: recombinant expression of a member of the ammonia monooxygenase superfamily. ISME J 6, 171–182 (2012). https://doi.org/10.1038/ismej.2011.98
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ismej.2011.98
Keywords
This article is cited by
-
Universal activity-based labeling method for ammonia- and alkane-oxidizing bacteria
The ISME Journal (2022)
-
Identification of active gaseous-alkane degraders at natural gas seeps
The ISME Journal (2022)
-
Verrucomicrobial methanotrophs grow on diverse C3 compounds and use a homolog of particulate methane monooxygenase to oxidize acetone
The ISME Journal (2021)
-
Research progress in bioremediation of petroleum pollution
Environmental Science and Pollution Research (2021)
-
Novel copper-containing membrane monooxygenases (CuMMOs) encoded by alkane-utilizing Betaproteobacteria
The ISME Journal (2020)