Abstract
The free-living but facultatively pathogenic amoebae of the genus Acanthamoeba are frequently infected with bacterial endosymbionts that can have a profound influence on the physiology and viability of their host. Parachlamydia acanthamoebae, a chlamydial endosymbiont in acanthamoebae, is known to be either symbiotic or lytic to its host, depending on the ambient conditions, for example, temperature. Moreover, parachlamydiae can also inhibit the encystment process in Acanthamoeba, an essential survival strategy of their host for the evasion of chemotherapeutic agents, heat, desiccation and radiation. To obtain a more detailed picture of the intracellular interactions of parachlamydiae and acanthamoebae, we studied parachlamydial infection in several Acanthamoeba isolates at the proteomic level by means of two-dimensional gel electrophoresis (2DE) and mass spectrometry. We observed that P. acanthamoebae can infect all three morphological subtypes of the genus Acanthamoeba and that the proteome pattern of released P. acanthamoebae elementary bodies was always practically identical regardless of the Acanthamoeba strain infected. Moreover, by comparing proteome patterns of encysting cells from infected and uninfected Acanthamoeba cultures, it was shown that encystment is blocked by P. acanthamoebae at a very early stage. Finally, on 2D-gels of purified P. acanthamoebae from culture supernatants, a subunit of the NADH-ubiquinone oxidoreductase complex, that is, an enzyme that has been described as an indicator for bacterial virulence was identified by a mass spectrometric and bioinformatic approach.
Similar content being viewed by others
Introduction
Free-living amoebae of the genus Acanthamoeba occur ubiquitously as predators of soil bacteria (Rodriguez-Zaragoza, 1994). Their life cycle consists of two stages, the feeding and multiplying trophozoite, and the rigid and endurable cyst, which displays resistance to antimicrobial agents, heat, desiccation, radiation and other environmental stresses. Besides their role as bacteriovores, acanthamoebae are also facultative human pathogens that can cause keratitis, mainly in contact lens wearers, and granulomatous encephalitis in immunocompromised individuals (Marciano-Cabral and Cabral, 2003). Moreover, Acanthamoeba and other free-living amoebae can act as vectors for bacteria that resist degradation after phagocytosis (Fritsche et al., 1993; Barker and Brown, 1994; Greub and Raoult, 2004). Indeed, as they were found to be associated with Legionella pneumophila (Rowbotham, 1980) and a number of other pathogenic bacteria such as Francisella tularensis (Abd et al., 2003) and Mycobacterium bovis (Taylor et al., 2003), free-living amoebae have been dubbed ‘trojan horses of the microbial world’ (Barker and Brown, 1994). Further studies on the vector role of free-living amoebae also revealed the existence of several previously unknown Chlamydia-like endosymbionts (Kahane et al., 1993; Amann et al., 1997; Rurangirwa et al., 1999; Horn et al., 2000; Collingro et al., 2005; Horn, 2008; Greub, 2009) whose potential as human pathogens is still being evaluated. Mainly respiratory tract infections have been suggested to be caused by these novel chlamydiae (Greub and Raoult, 2002a; Friedman et al., 2003; Haider et al., 2008) but also certain evidence for a possible role in adverse pregnancy outcomes exists (Baud et al., 2007). It could also be of medical relevance that the presence of Chlamydia-like endosymbionts enhances the cytopathogenicity of Acanthamoeba to human cells (Fritsche et al., 1998). However, no data on the influence of bacterial endosymbionts on the course of Acanthamoeba keratitis, or granulomatous Acanthamoeba encephalitis respectively, are currently available.
One of these Chlamydia-like bacteria, Parachlamydia acanthamoebae, has been reported to have a profound effect on the physiology of its Acanthamoeba host. Depending on the temperature, P. acanthamoebae can be endosymbiotic (25–30 °C) or lytic (32–37 °C) for Acanthamoeba (Greub et al., 2003a). Moreover, infected Acanthamoeba strains lose their ability to form cysts (Michel et al., 2001), which can be highly disadvantageous because encystment constitutes Acanthamoeba's main survival strategy for the evasion of adverse environmental conditions. As is the case with the human pathogens Chlamydophila pneumoniae and Chlamydia trachomatis, P. acanthamoebae is an obligate intracellular parasite, which is almost exclusively confined to vacuoles, termed inclusions, where it proliferates. The dividing stage, that is, the vulnerable reticulate body, which lacks a thick cell wall and which is only found within this vacuole (Amann et al., 1997), finally transforms into the more rigid elementary body, which stains Gram-positive. The latter is the infectious stage, which accumulates in the vacuole until the host cell is lysed, thereby liberating the elementary bodies, which can then start another round of infection. Alternatively, the bacteria can also be expelled from the cell in a vesicle (Greub and Raoult, 2002b). In contrast to C. pneumoniae and C. trachomatis, however, P. acanthamoebae has an astonishingly wide host range and cannot only infect Acanthamoeba but also distantly related or even unrelated free-living amoebae like Balamuthia mandrillaris, Hartmannella vermiformis and Naegleria spp. (Michel et al., 2001). Also human cells, such as macrophages, lung fibroblasts or pneumocytes can be infected by P. acanthamoebae (Greub et al., 2003b; Casson et al., 2006), raising serious concerns about P. acanthamoebae as a human pathogen (Haider et al., 2008; Horn, 2008; Greub, 2009).
Despite its presumptive ecological and medical importance, relatively little is known about P. acanthamoebae infections at the molecular level. In this study, we took advantage of a P. acanthamoebae strain, which was accidently introduced to our laboratory and used it to shed more light on the interactions of P. acanthamoebae and its Acanthamoeba host at the molecular level. It was our aim to investigate the phenomenology of P. acanthamoebae infection in its Acanthamoeba hosts by application of two-dimensional gel electrophoresis (2DE). Infected Acanthamoeba cultures as well as purified P. acanthamoebae elementary bodies were studied and P. acanthamoebae's capability to abrogate encystment in Acanthamoeba was investigated.
Materials and methods
Cell culture
All Acanthamoeba strains used in this study were grown as described (Köhsler et al., 2008). Strain PAT06, group II, genotype T4, was isolated from a keratitis patient in 2006 and strain NEFF, group II, genotype T4 (ATCC 30010), was obtained from the American Type Culture Collection. Strain 72/2, group III, genotype T5 (ATCC 50704), had been originally isolated from human nasal mucosa, and strain PB30/40, group I, genotype T7 from a greenhouse (Michel et al., 2004).
Escherichia coli XL-1 Blue was grown at 37 °C in liquid lysogeny broth medium containing 2.5 μg ml−1 tetracyclin.
Cocultivation of P. acanthamoeba in Acanthamoeba strains
Infected cultures were grown at room temperature and subcultured every 3 or 4 days. When Acanthamoeba cultures were purposely infected, a small amount (approximately 200 μl) of supernatant of an infected Acanthamoeba culture containing P. acanthamoebae elementary bodies was added to non-infected cultures. Acanthamoeba cells from the already infected culture had been previously removed by centrifugation at low g-force (600 g).
Induction of encystment
Encystment was induced in Tris-buffered starvation medium as described in Köhsler et al. (2008).
Cell fractionation
After harvest at 800 g (5 min) infected or uninfected cells were washed twice in 1 × PBS. Afterwards, cells were resuspended in a buffer (SMD) containing 250 mM sucrose, 10 mM MOPS (3-morpholinopropane-1-sulfonic acid) pH 7.2, and 10 mM DTT (dithiothreitol) and broken up in a Dounce homogenizer. After removal of cell debris (1000 g, 3 min) the cell lysate was centrifuged at 20 000 g for 20 min to obtain the large organelle fraction and intact parachlamydiae. Pellets were resuspended in SMD buffer and the suspension constituents were further separated by centrifugation over a 45% Percoll gradient in SMD (100 000 g, 1 h). All centrifugation steps were performed at 4 °C.
Two-dimensional gel electrophoresis
Sample preparation of cells for two-dimensional gel electrophoresis (2DE) and 2DE were performed as described (Leitsch et al., 2005). Gel images were evaluated using Melanie 4 software (Genebio, Geneva, Switzerland).
Mass spectrometry
Spots of interest were manually excised for MS analysis from Coomassie-stained gels. Peptide mass fingerprinting (PMF) and post source decay (PSD) experiments were performed on a MALDI-reflectron Time-of-flight instrument (AXIMA-CFRplus, Shimadzu Biotech Kratos Analytical, Vienna, Austria). External calibration was performed based on monoisotopic values of well-defined peptides. Obtained mass spectra were corrected for matrix- and enzyme-derived interferences (Keller et al., 2008). The resulting monoisotopic list of m/z values was submitted to the search engine MASCOT (Perkins et al., 1999) searching all proteins and DNA sequence information from public databases (MSDB, Swiss-Prot, NCBInr) and the translated partial genome sequence information from Parachlamydia acanthamoebae UV7 and Acanthamoeba castellanii containing 2931 Parachlamydia sequences and 12 990 sequences of Acanthamoeba. The following search parameters were applied: mass accuracy, ±0.35 Da, fixed modification—carbamidomethylation, variable modifications—methionine oxidation, missed cleavages: and a maximum of one tryptic missed cleavage. Based on the measured PMF peptides were selected for seamless PSD experiments. Search parameters for PSD experiments were identical to PMF experiments, except for peptide mass accuracy, ±1 Da, product ion tolerance, ±1.0 Da. A protein was considered as identified, if the scores of database searches clearly exceeded the algorithm's significance threshold (P<0.05) for PMF and PSD data.
Identification of Parachlamydia acanthamoebae
Simultaneous isolation of DNA from the Acanthamoeba host and their bacterial endosymbionts was performed using the DNeasy blood & tissue kit (Qiagen, Hilden, Germany) according to the protocol recommended by the manufacturer. Oligonucleotide primers panF (5′-CGTGGATGAGGCATGCRAGTCG-3′) and panR (5′-GTCATCRGCCYYACCTTVSRCRYYTCT-3′) targeting most chlamydiae (Corsaro et al., 2002) were used for PCR to obtain near full-length bacterial 16S rRNA gene fragments, which were sequenced directly on an ABI 3130 XL genetic analyzer using the BigDye Terminator kit v3.1 (Applied Biosystems, Vienna, Austria).
For fluorescence in situ hybridization, amoebae were harvested by centrifugation (4000 g, 5 min) and washed with 1 × Page's Amoebic Saline (0.12 g l−1 NaCl, 0.004 g l−1 MgSO4 × 7H2O, 0.004 g l−1 CaCl2 × 2H2O, 0.142 g l−1 Na2HPO4, 0.136 g l−1 KH2PO4). After resuspension in 1 × Page's Amoebic Saline, amoebae were allowed to attach to the surface of glass slides for 20 min. Cells were fixed with 4% paraformaldehyde for 10 min at room temperature. Hybridization was performed using the protocol, hybridization and washing buffer described previously (Daims et al., 2005). The oligonucleotide probes Euk516-Fluos, targeting eukaryotic 18S rRNA (Amann et al., 1990), and Bn9-658-Cy3, targeting 16S rRNA of parachlamydiaceae (Poppert et al., 2002) were used. Hybridized slides were examined using a confocal laser scanning microscope (LSM 510 Meta, Carl Zeiss, Vienna, Austria) equipped with two helium-neon-lasers (543 and 633 nm) and an argon laser (458–514 nm). Image analysis was performed with the standard software package delivered with the instrument (version 3.2).
Strain accession numbers
Strain PAT06 was deposited at the ATCC and is available under the following accession number NRS-10465 (preliminary number).
Results
Identification of a highly virulent Parachlamydia acanthamoebae strain that efficiently infects all three morphological subtypes of Acanthamoeba
Around the end of 2007, we observed the presence of intracellular bacteria in several Acanthamoeba strains that are grown in our laboratory. These bacteria visibly proliferated in vacuoles and finally lysed their host (Figure 1a) as typically observed with P. acanthamoebae (Horn, 2008). After lysis, the bacteria were released into the surrounding growth medium. By applying fluorescence in situ hybridization (Figure 1b) and sequencing of the 16S rRNA gene, the endosymbiont was indeed identified as Parachlamydia acanthamoebae with the highest sequence similarity to Parachlamydia acanthamoebae strain Seine (GenBank: DQ309029). In accordance with previous observations on P. acanthamoebae (Michel et al., 2001), our strain displayed a wide host range and could infect Acanthamoeba strains from all three morphological subtypes, PB 30/40, NEFF, and 72/2 (Table 1).
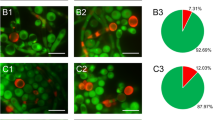
Images showing A. castellanii NEFF infected with P. acanthamoebae. (a), Stages of infection with P. acanthamoebae in Acanthamoeba castellanii (NEFF strain). The bacteria proliferate in inclusions (1) until the whole cell is totally crammed (2). Finally, the host cells rupture and the bacteria are released into the surrounding growth medium (3). Arrows mark inclusions. Photographs were taken with a Nikon Digital Sight DS-L1 camera on a Nikon Labophot-2 microscope. (b), Fluorescence in situ hybridization image showing an A. castellani NEFF cell, which is heavily infected with P. acanthamoebae. Host cells were stained with the probe Euk516-Fluos (green), bacteria with the probe Bn9-658-Cy3 (red). The parachlamydiae are confined to inclusions. The bar equals 5 μm.
2DE images of infected Acanthamoeba cells
We applied two-dimensional gel electrophoresis (2DE) to visualize differences in the proteomic patterns of infected and uninfected acanthamoebae. Figure 2 shows the 2DE images of a recently isolated, pathogenic strain belonging to genotype T4 (morphological type II), PAT06, either uninfected or infected with P. acanthamoebae. In the 2DE images of infected cells, numerous highly expressed proteins were detected, which were absent from the 2DE images of uninfected cells. Identical changes with regard to protein content were also observed in the 2D-gels of other infected Acanthamoeba species (Figure 3a): A. castellanii (NEFF), A. comandoni (PB30/40) and A. lenticulata (72/2).
The 2DE images of infected Acanthamoeba PAT06 cells dramatically differ from those of uninfected cells. A number of highly expressed new proteins appear (encircled) which are also visible on Coomassie-stained 2D-gels of P. acanthamoebae elementary bodies isolated from the supernatant of a PAT06 culture (right). Two identified proteins of P. acanthamoebae: NADH-ubiquinone oxidoreductase D subunit (1) and elongation factor Tu (2). The respective protein spots are marked on the 2D-gels of infected PAT06 cells (center) and of purified P. acanthamoebae (right). Gels were stained with Coomassie Brilliant Blue. The gel images can be seen in larger format in Supplementary Figure 1.
All three morphological subtypes of Acanthamoeba can host P. acanthamoebae. (a), the same changes as observed in PAT06 are found in infected cells of three other strains, each representing another morphological type of the genus Acanthamoeba. (b), P. acanthamoebae isolated from the supernatants of PAT06 (left), NEFF (middle) and 72/2 cultures (right) display practically identical 2DE patterns. Gels were stained with Coomassie Brilliant Blue. The gel images can be seen in a larger format in Supplementary Figure 2.
The bacterial origin of the novel proteins was confirmed by performing 2DE with purified P. acanthamoebae elementary bodies from culture supernatants of an infected PAT06 culture (Figure 2). A comparably small total number of only 426 spots was detected on Coomassie-stained 2D-gels (pH-range 3–10) of purified P. acanthamoebae when using 2DE-imaging software (Melanie 4, Genebio) (Figure 2). Remarkably, the 10 most prominent protein spots accounted for more than 40% of the total protein content as judged by densitometrical analysis with our 2DE-imaging software. For comparison, 2DE was also performed with extracts of Escherichia coli XL-1 Blue by use of the same protocol (Supplementary Figure 1), which resulted in approximately the double number of proteins visualized (847). In contrast to P. acanthamoebae, the 10 most prominent spots accounted for only 17% of the total protein content visualized.
We isolated a selection of nine spots from 2D-gels of purified P. acanthamoebae and analysed them by MALDI mass spectrometry. Unfortunately, either due to the still incomplete genome data of P. acanthamoebae strain UV7 or due to strain differences, only two proteins could be identified with statistical significance (Figure 2, Supplementary Table 1): Na(+)-translocating NADH-ubiquinone oxidoreductase subunit D (spot 1) and elongation factor Tu (spot 2). Spot 1 containing NADH-ubiquinone oxidoreductase subunit D was by far the most prominent spot on the 2D-gel of the P. acanthamoebae isolate, comprising more than 15% of the total protein content. However, it also overlapped with at least one, yet unidentified, protein with only slightly different pI and molecular weight.
The protein content of P. acanthamoebae isolated from culture supernatants is widely identical regardless of the Acanthamoeba species parasitized
We isolated P. acanthamoebae elementary bodies from the culture supernatants of three different Acanthamoeba species, or strains respectively, that is, PAT06, NEFF and 72/2, to investigate by 2DE whether the protein composition of the parachlamydiae differs depending on the host parasitized. One rationale for this experiment was a report on coating of the outer membrane of L. pneumophila with acanthamoebal proteins after lysis of the Acanthamoeba host (Barker et al., 1993). Accordingly, we wanted to check whether a similar effect occurs with P. acanthamoebae.
However, we did not find any evidence for an influence of the host on the protein composition of P. acanthamoebae, the proteomic images of the isolated parachlamydiae all being practically identical (Figure 3b). The minute differences visible are due to methodological constraints of 2DE (especially at the borders of the gel) and due to slightly different amounts of protein loaded. This result does rather argue against any coating of P. acanthamoebae with host protein because if Acanthamoeba proteins had been introduced to the parachlamydiae, they would have most probably located to different positions on the 2D-gels of the P. acanthamoebae extracts due to their strain or species-specific differences with regard to pI and molecular mass. Indeed, the proteomic patterns of the three Acanthamoeba strains are strongly divergent (Figures 3a and b, Supplementary Figure 2) and hardly any protein spot in the 2DE image of A. lenticulata 72/2 can be matched to A. castellanii PAT06 or NEFF, thereby underlining the high genetic diversity within the genus Acanthamoeba.
None of the previously detected parachlamydial proteins localize to the cytosolic compartment of the Acanthamoeba host
We wanted to investigate whether the bacterial proteins detected in infected PAT06 cells (Figure 2) localize to different compartments of the host cell. To this end, we performed a differential centrifugation experiment with lysates of infected and uninfected PAT06 cells (Supplementary Figure 3). Interestingly, none of the parachlamydial proteins, previously observed in 2D-gels from purified P. acanthamoebae could be found on 2D-gels of the 20 000 g supernatant fraction of infected PAT06 (Figure 4a). This fraction mainly consists of cytosolic proteins. Moreover, the 2DE images of the 20 000 g supernatant fraction from infected and uninfected PAT06 (Supplementary Figure 3) were almost identical with only a small number of host proteins obviously differentially regulated (Figure 4b). One spot in the acidic, low molecular mass range was found to be present in 2D-gels of the 20 000 g supernatant-fraction from infected cells while being absent in the respective fraction of uninfected PAT06 (Supplementary Figure 3). But as we have observed this spot in earlier experiments also on 2D-gels from uninfected cells (not shown), it was not considered for further analysis.
No proteins previously detected on 2D-gels of P. acanthamoebae elementary bodies were detected in the cytosol of the host. (a), All prominent proteins seen on 2D-gels of P. acanthamoebae (right) are also present on 2D-gels of the high density band, which is obtained after running the easily precipitable constituents of infected PAT06 cells (20 000 g, 20 min) over a 45% Percoll gradient (middle). The gel images can be seen in larger format in Supplementary Figure 3. (b) Two sections of 2D-gels from the 20 000 g lysate supernatant of infected and non-infected PAT06 cells showing differentially expressed proteins. Expression levels are given as percentages of total protein visualized. Gels were stained with Coomassie Brilliant Blue. Image analysis was performed with Melanie 4 software.
All parachlamydial proteins, previously detected in 2D-gels from purified P. acanthamoebae and from infected Acanthamoeba cells (Figures 2 and 3), were observed on 2D-gels of the high density fraction of a Percoll gradient from the 20 000 g precipitable pellet of infected PAT06 (Figure 4a). This fraction mainly consisted of intact parachlamydiae. Interestingly, only 15 spots were detected that did not match to a corresponding spot in 2D-gels of purified P. acanthamoebae. Of these, however, merely one spot matched to a spot in a 2D-gel of the high density fraction of uninfected cells (Supplementary Figure 3). Thus, it is possible that the remaining 14 spots also contain proteins of parachlamydial origin which, however, are only expressed in the reticulate body stage.
Parachlamydia acanthamoebae inhibits the encystment process of Acanthamoeba in an early stage
Besides being lytic to Acanthamoeba at temperatures above 32 °C (Greub et al., 2003a), P. acanthamoebae also has a strong impact on the life cycle of its host by inhibiting the encystment process (Michel et al., 2001). We reasoned that the arrest of encystment would occur at a certain stage, which could be visualized by applying 2DE and therefore performed an encystment experiment in infected and uninfected PAT06 cells. Strain PAT06 was chosen because it was a fresh isolate that had so far retained its ability to encyst whereas older isolates usually display strongly reduced encystment rates (Köhsler et al., 2008). When we induced encystment with infected PAT06 cells by using a Tris-buffered starvation medium (Hirukawa et al., 1998; Köhsler et al., 2008), no mature cysts were formed. Most of the cells in the culture were lysed within 24 h but a proportion of infected cells survived extended periods of time in starvation medium (at least 3 days). The 2DE images of these survivors displayed P. acanthamoebae proteins but were devoid of the most conspicuous spots of 2D-gels of normal cysts (Figure 5). Interestingly, two cleavage products of actin, which appear already shortly after induction of encystment (less than 8 h) and, which persist during the process (Leitsch et al., 2010) were absent in the survivors of the infected culture (Figure 5). This result suggests that P. acanthamoebae interferes with the encystment of its host at a very early stage. Finally, we observed that although parachlamydial proteins were readily observable on 2D-gels of infected cells that had survived in starvation medium, the largest spot, containing NADH-ubiquinone oxidoreductase D subunit was only very faint (Figure 5).
P. acanthamoebae abrogates encytment in Acanthamoeba castellanii in an early stage. 2D-gels of infected PAT06 cultures of cells that survived after induction of encystment (left) lack two actin fragments (encircled, full line) that are already present in encysting cells, 8 h after placement in starvation medium (right). The surviving cells of infected cultures comprise a number of parachlamydial proteins (encircled, dashed line) but widely lack NADH-ubiquinone oxidoreductase D subunit. Gels were stained with Coomassie Brilliant Blue. Image analysis was performed with Melanie 4 software. The gel images can be seen in a larger format in Supplementary Figure 4.
Discussion
We studied several aspects of P. acanthamoebae infections in Acanthamoeba by applying 2DE. Based on our data, a number of conclusions can be drawn. First, the visualized protein content of P. acanthamoebae after lysis of its host was always practically identical, independent of the Acanthamoeba species parasitized (Figure 3b). This rather argues against any specific adaptations to different hosts in terms of alterations in parachlamydial gene expression. Furthermore, in contrast to observations made with L. pneumophila (Barker et al., 1993), we could not find any evidence for coating of P. acanthamoebae with detectable amounts of host protein (Figure 3b).
Second, the proportion of P. acanthamoebae proteins that enter the cytosol of the host must be small because we could not find any proteins previously identified as being of parachlamydial origin on 2D-gels of the cytosolic fraction of infected Acanthamoeba cells (Figure 4a). Of course, as 2DE only visualizes intermediately to highly expressed proteins, low copy proteins are easily overlooked. Thus, the transport of parachlamydial low copy effector proteins into the host's cytoplasm cannot be ruled out. In fact, human pathogens such as C. pneumoniae and C. trachomatis ingeniously manipulate the host by trafficking various effectors out of the inclusion into the host cytoplasm. These factors prevent apoptosis, MHC expression, and lipid antigen presentation (Valdivia, 2008). However, in human macrophages, P. acanthamoebae cannot prevent induction of apoptosis (Greub et al., 2003b) and studies that show such subtle manipulation strategies in P. acanthamoebae are still missing. Nevertheless, although no parachlamydial proteins were detected in the cytosolic fraction of infected amaoebae, we did identify a small number of host proteins to be differentially expressed (Figure 4b) in infected and uninfected cells. As yet, the significance of this finding remains unclear and the identification of the host proteins affected is required.
Third, as compared with, for example, E. coli, the proteome of P. acanthamoebae elementary bodies comprises only a comparably small number of proteins, that is, 426 protein spots when 2D-gels were stained with Coomassie Brilliant Blue (Figure 2). Strikingly, the 10 most prominent spots in 2D-gels from P. acanthamoebae account for as much as 40% of the protein content in the gels, whereas the 10 most prominent spots in 2D-gels from E. coli XL-1 Blue account only for 17% of the protein content. The most prominent spot on 2D-gels of P. acanthamoebae elementary bodies contained Na(+)-translocating NADH-ubiquinone oxidoreductase D subunit (Figure 2, Supplementary Table 1). However, the protein spot also contained at least one other protein which, unfortunately, could not be identified. Thus, at present, it is not possible to make any conclusive predictions with regard to this protein's expression level. Still, as NADH-ubiquinone oxidoreductase has been suggested to be indicative of bacterial virulence (Häse et al., 2001) and as the exploitation of Na+ gradients in energy metabolism has been suggested to be a widespread strategy among human pathogens, including C. pneumoniae and C. trachomatis (Häse et al., 2001), the potentially very high expression of this enzyme in P. acanthamoebae warrants further investigation.
Finally, parachlamydiae obviously block the encystment process in the Acanthamoeba host at a very early stage (Figure 5). Two actin isoforms, which are generated by the proteolytic cleavage of actin and which appear earlier than 8 h after induction of encystment in uninfected cells (Leitsch et al., 2010), were completely absent in infected Acanthamoeba PAT06 cultures, even after prolonged incubation (at least 3 days). Possibly, the proliferating parachlamydiae deprive their Acanthamoeba host of the resources that are needed for entering the encystment process. Alternatively, parachlamydiae could deactivate cysteine proteinases or prevent their release. Cysteine proteinases have a very important role during the early phase of encystment in A. castellanii and seem to be confined to vesicles rather than being cytosolic (Leitsch et al., 2010). Thus, it is conceivable that parachlamydiae and cysteine proteinases localize to the same intracellular compartment. Of course, as discussed above, encystment could also be blocked by parachlamydial effector proteins, which are expressed at levels low enough to be overlooked on 2D-gels. In any case, it is probable that, very much like shifting infected Acanthamoeba cultures to temperatures higher than 32 °C (Greub et al., 2003a), induction of encystment signals P. acanthamoebae to start rapid proliferation and to lyse its host. Interestingly, however, not all thus treated Acanthamoeba cells were lysed. On the 2DE images of surviving cells P. acanthamoebae proteins were observed, but the most prominent spot, containing the D-subunit of NADH-ubiquinone oxidoreductase, was widely missing. The significance of this observation will be addressed in our future work.
Accession codes
References
Abd H, Johansson T, Golovliov I, Sandström G, Forsman M . (2003). Survival and growth of Francisella tularensis in Acanthamoeba castellanii. Appl Environ Microbiol 69: 600–606.
Amann R, Springer N, Schönhuber W, Ludwig W, Schmid EN, Müller K et al. (1997). Obligate intracellular bacterial parasites of acanthamoebae related to Chlamydia spp. Appl Environ Microbiol 63: 115–121.
Amann RI, Binder BJ, Olson RJ, Chisholm SW, Devereux R, Stahl DA . (1990). Combination of 16S rRNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Appl Environ Microbiol 56: 1919–1925.
Barker J, Brown MRW . (1994). Trojan horses of the microbial world: protozoa and the survival of bacterial pathogens in the environment. Microbiology 240: 1253–1259.
Barker J, Lambert PA, Brown MRW . (1993). Influence of intra-amoebic and other growth conditions on the surface properties of Legionella pneumophila. Infect Immun 61: 3503–3510.
Baud D, Thomas V, Arafa A, Regan L, Greub G . (2007). Waddlia chondrophila, a potential agent of human fetal death. Emerg Infect Dis 13: 1239–1243.
Casson N, Medico N, Bille J, Greub G . (2006). Parachlamydia acanthamoebae enters and multiplies within pneumocytes and lung fibroblasts. Microb Infect 8: 1294–1300.
Collingro A, Poppert S, Heinz E, Schmitz-Esser S, Essig A, Schweikert M et al. (2005). Recovery of an environmental Chlamydia strain from activated sludge by co-cultivation with Acanthamoeba spp. Microbiology 151: 301–309.
Corsaro D, Venditti D, Valassina M . (2002). New parachlamydial 16S rDNA phylotypes detected in human clinical samples. Res Microbiol 153: 563–567.
Daims H, Stoecker K, Wagner M . (2005). Fluorescence in situ hybridization for the detection of prokaryotes. In Osborn AM, Smith CJ (eds). Advanced Methods in Molecular Microbial Ecology. Bios-Garland: Abingdon, UK, pp. 213–239.
Friedman MG, Dvoskin B, Kahane S . (2003). Infections with the chlamydia-like microorganism Simkania negevensis, a possible emerging pathogen. Microbes Infect 5: 1013–1021.
Fritsche TR, Gautom RK, Seyedirashti S, Bergeron DL, Lindquist TD . (1993). Occurrence of bacterial endosymbionts in Acanthamoeba spp. isolated from corneal and environmental specimens and contact lenses. J Clin Microbiol 31: 1122–1126.
Fritsche TR, Sobek D, Gautom RK . (1998). Enhancement of in vitro cytopathogenicity by Acanthamoeba spp. following acquisition of bacterial endosymbionts. FEMS Microbiol Lett 166: 231–236.
Greub G . (2009). Parachlamydia acanthamoebae, an emerging agent of pneumonia. Clin Microbiol Infect 15: 18–28.
Greub G, La Scola B, Raoult D . (2003a). Parachlamydia acanthamoeba is endosymbiotic or lytic for Acanthamoeba polyphaga depending on the incubation temperature. Ann NY Acad Sci 990: 628–634.
Greub G, Mege J, Raoult D . (2003b). Parachlamydia acanthamoeba enters and multiplies within human macrophages and induces their apoptosis. Infect Immun 71: 5979–5985.
Greub G, Raoult D . (2002a). Parachlamydiaceae: potential emerging pathogens. Emerg Infect Dis 8: 625–630.
Greub G, Raoult D . (2002b). Crescent bodies of Parachlamydia acanthamoeba and its life cycle within Acanthamoeba polyphaga: an electron micrograph study. Appl Environ Microbiol 68: 3076–3084.
Greub G, Raoult D . (2004). Microorganisms resistant to free-living amoebae. Clin Microbiol Rev 17: 413–433.
Haider S, Collingro A, Walochnik J, Wagner M, Horn M . (2008). Chlamydia-like bacteria in respiratory samples of community-acquired pneumonia patients. FEMS Microbiol Lett 281: 198–202.
Häse C, Fedorova ND, Galperin MY, Dibrov PA . (2001). Sodium ion cycle in bacterial pathogens: evidence from cross-genome comparisons. Microbiol Mol Biol Rev 65: 353–370.
Hirukawa Y, Nakato H, Izumi S, Tsuruhara T, Tomino S . (1998). Structure and expression of a cyst specific protein of Acanthamoeba castellanii. Biochim Biophys Acta 1398: 47–56.
Horn M . (2008). Chlamydiae as symbionts in eukaryotes. Ann Rev Microbiol 62: 113–131.
Horn M, Wagner M, Müller K, Schmid EN, Fritsche TR, Schleifer K et al. (2000). Neochlamydia hartmannellae gen. nov., sp. nov. (Parachlamydiaceae), an endoparasite of the amoeba Hartmannella vermiformis. Microbiology 146: 1231–1239.
Kahane S, Gonen R, Sayada C, Elion J, Friedman MG . (1993). Description and partial characterization of a new Chlamydia-like microorganism. FEMS Microbiol Lett 109: 329–333.
Keller BO, Sui J, Young AB, Whittal RM . (2008). Interferences and contaminants encountered in modern mass spectrometry. Anal Chim Acta 627: 71–81.
Köhsler M, Leitsch D, Fürnkranz U, Duchêne M, Aspöck H, Walochnik J . (2008). Acanthamoeba strains lose their abilities to encyst synchronously upon prolonged axenic culture. Parasitol Res 102: 1069–1072.
Leitsch D, Köhsler M, Marchetti-Deschmann M, Deutsch A, Allmaier G, Duchêne M et al. (2010). A major role for cysteine proteases during the early phase of Acanthamoeba castellanii encystment. Eukaryot Cell 9: 611–618.
Leitsch D, Radauer C, Paschinger K, Wilson IBH, Breiteneder H, Scheiner O et al. (2005). Entamoeba histolytica: analysis of the trophozoite proteome by two-dimensional polyacrylamide gel electrophoresis. Exp Parasitol 110: 191–195.
Marciano-Cabral F, Cabral G . (2003). Acanthamoeba spp. as agents of disease in humans. Clin Microbiol Rev 16: 273–307.
Michel R, Müller KD, Hoffmann R . (2001). Enlarged Chlamydia-like organisms as spontaneous infection of Acanthamoeba castellanii. Parasitol Res 87: 248–251.
Michel R, Steinert M, Zeller L, Hauröder B, Henning K . (2004). Free-living amoebae may serve as hosts fort the chlamydia-like bacterium Waddlia chondrophila isolated from an aborted bovine foetus. Acta Protozool 43: 37–42.
Perkins DN, Pappin DJ, Creasy DM, Cottrell JS . (1999). Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis 20: 3551–3567.
Poppert S, Essig A, Marre R, Wagner M, Horn M . (2002). Detection and differentiation of chlamydiae by fluorescence in situ hybridization (FISH). Appl Environ Microbiol 68: 4081–4089.
Pussard M, Pons R . (1977). Morphologie de la paroi kystique et taxonomie du genre Acanthamoeba (Protozoa, Amoebida). Protistologica 13: 557–598.
Rodriguez-Zaragoza S . (1994). Ecology of free-living amoebae. Crit Rev Microbiol 20: 225–241.
Rowbotham TJ . (1980). Preliminary report on the pathogenicity of Legionella pneumophila for freshwater and soil amoebae. J Clin Pathol 33: 1179–1183.
Rurangirwa FR, Dilbeck PM, Crawford TB, McGuire TC, McElwain TF . (1999). Analysis of the 16S rRNA gene of microorganism WSU 86-1044 from an aborted bovine foetus reveals that it is a member of the order Chlamydiales: proposal of Waddliaceae fam. nov., Waddlia chondrophila gen. nov., sp. nov. Int J Syst Bacteriol 49: 577–581.
Taylor SJ, Ahonen LJ, de Leij FAAM, Dale JW . (2003). Infection of Acanthamoeba castellanii with Mycobacterium bovis and M. bovis BCG and survival of M. bovis within the amoebae. Appl Environ Microbiol 69: 4316–4319.
Valdivia RH . (2008). Chlamydia effector proteins and new insights into chlamydial cellular microbiology. Curr Opin Microbiol 11: 53–59.
Acknowledgements
This work was supported by Grants P19044 and Y277-B03 of the Austrian Science Fund (FWF). We are especially grateful to Matthias Horn for providing us with his Parachlamydia acanthamoebae UV7 genome data base and for his critical reading of the article.
Author information
Authors and Affiliations
Corresponding author
Additional information
Supplementary Information accompanies the paper on The ISME Journal website
Rights and permissions
About this article
Cite this article
Leitsch, D., Köhsler, M., Marchetti-Deschmann, M. et al. Proteomic aspects of Parachlamydia acanthamoebae infection in Acanthamoeba spp.. ISME J 4, 1366–1374 (2010). https://doi.org/10.1038/ismej.2010.68
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ismej.2010.68
Keywords
This article is cited by
-
An unusual thioredoxin system in the facultative parasite Acanthamoeba castellanii
Cellular and Molecular Life Sciences (2021)
-
‘Candidatus Cochliophilus cryoturris’ (Coxiellaceae), a symbiont of the testate amoeba Cochliopodium minus
Scientific Reports (2017)
-
Legionella pneumophila prevents proliferation of its natural host Acanthamoeba castellanii
Scientific Reports (2016)