Abstract
In the present study, we tested the hypothesis that animal treading associated with a high input of organic matter would favour methanogenesis in soils used as overwintering pasture. Hence, methane emissions and methanogen populations were examined at sections with different degree of cattle impact in a Farm in South Bohemia, Czech Republic. In spring, methane emission positively corresponded to the gradient of animal impact. Applying phospholipid etherlipid analysis, the highest archaeal biomass was found in section severe impact (SI), followed by moderate impact (MI) and no impact. The same trend was observed for the methanogens as showed by real-time quantitative PCR analyses of methyl coenzyme M reductase (mcrA) genes. The detection of monounsaturated isoprenoid side chain hydrocarbons (i20:1) indicated the presence of acetoclastic methanogens in the cattle-impacted sites. This result was corroborated by the phylogenetic analysis of mcrA gene sequences obtained from section SI, which showed that 33% of the analysed clones belonged to the genus Methanosarcina. The majority of the sequenced clones (41%) showed close affiliations with uncultured rumen archaeons. This leads to the assumption that a substantial part of the methanogenic community in plot SI derived from the grazing cattle itself. Compared to the spring sampling, in autumn, a significant reduction in archaeal biomass and number of copies of mcrA genes was observed mainly for section MI. It can be concluded that after 5 months without cattle impact, the severely impact section maintained its methane production potential, whereas the methane production potential under moderate impact returned to background values.
Similar content being viewed by others
Introduction
Agriculture offers a great potential for green house gas mitigation and for a systematic sequestration of carbon dioxide in soils and in plant biomass (OCA, 2006). In this respect, upland grasslands in low-input farming systems are considered as sinks for the green house gas methane (CH4) (Hütsch et al., 1994) and only weak sources for nitrous oxide (N2O) (Mosier et al., 1991). However, these mitigation potential can change when grasslands are used as pastures by grazing cattle. For example, Clayton et al. (1994) found that N2O emissions from a grazed pasture were three times higher than those from the pasture without grazing and speculated that interaction between treading and excretal returns could have stimulated this process. Animal treading causes reduction in soil aeration through soil compaction, which may enhance rates of denitrification (Menneer et al., 2005). Moreover, treading can also mix organic carbon-rich material originated from excrement into soil, stimulating microbial metabolism and thus increasing oxygen demand also at lower soil depths (Šimek et al., 2006). Both, anaerobic environment and availability of organic carbon may favour methanogenesis. Indeed, some studies have shown a temporal increase in methane emissions immediately after application of manure for soil fertilization (Sommer et al., 1996; Sherlock et al., 2002).
Although changes in methane fluxes are expected in soils strongly impacted by animal husbandry, data about the structure and function of methanogenic archaeal community present in this environment are missing. Therefore, the objective of this study was to determine the effect of outdoor overwintering cattle husbandry on methane emissions in coherency to the abundance and diversity of soil methanogenic communities. Particularly, the following hypothesis were tested: (i) cattle overwintering pasture leads to changes in soil function, towards methane emission instead of methane sink, (ii) this emissions are correlated with the degree of animal impact and (iii) cattle excrements serve as an inoculum for rumen methanogens and stimulate growth and activity of soilborne methanogens.
To characterize total and methanogenic archaeal biomass, we used the phospholipid etherlipid (PLEL) approach (Gattinger et al., 2002, 2003). The taxonomic resolution of this method is relatively low, but its robustness towards varying soil matrices has been demonstrated (Bai et al., 2000; Wachinger et al., 2000; Wagner et al., 2005). Moreover, real-time quantitative PCR and clone library analyses of the methyl coenzyme M reductase (mcrA) gene, which codes for methyl-coenzyme M reductase α-subunit, were conducted for more detailed investigation of the abundance and structure of methanogenic communities. This enzyme catalyses the reduction of methyl-coenzyme M leading to the release of methane (Ellermann et al., 1988). The presence of the mcrA gene is restricted to methanogenic archaea (Thauer, 1998); hence its quantity serves as an estimate for methanogenic biomass in soil.
In the present study, we demonstrated that use of grasslands for cattle overwintering drastically changed soil properties, which in turn affected the abundance and activity of soil methanogens.
Material and methods
Experimental site
Investigations were carried out at a cattle overwintering area located at Borová Farm near Český Krumlov in South Bohemia, Czech Republic (latitude 48°52′ N, longitude 14°13′ E). The approximately 4-ha area has been used for overwintering (from October/November until April/May) of about 90 cows since 1995 (for more details see Šimek et al., 2006). The soil on the site is a sandy loam, classified as Cambisol, and containing 60–80% sand, 14–32% silt and 6–14% clay (according to USDA classification system). The plant cover is a perennial mixture of grasses, clovers and other dicotyledonous plants. The long-term mean annual precipitation in the area is 650 mm and the annual average temperature is 7°C (data from meteorological station located at 7 km distance from the experimental farm). At the end of the winter season (from April to May), there is a visible gradient of impact due to the cattle husbandry. The degree of impact on overwintering sites is not identical on the whole pasture. We investigated three sections of this gradient: the most affected area situated near the barn (section SI=severe impact), a less impacted area in the middle of the gradient (section MI=moderate impact) and unaffected area at the opposite side of the overwintering area (section NI=no impact). In spring, the studied sections were characterized as follows: SI by completely destroyed vegetation, damaged soil structure and oversaturation by the excrements; MI by partly destroyed vegetation and remains of excrements on the soil surface; NI by undisturbed grass cover and no visible remains of excrements.
Methane flux measurements and soil sampling
Field works were performed twice, in spring (11 May 2005), after removing the animals from overwintering area and in fall (18 October 2005), before they were brought back. Different locations at each section were randomly selected from each section for gas flux measurements and soil sampling and used as independent field replicates in this study. Methane fluxes were determined using medium-sized (basal area, 0.076 m2; volume, 15 dm3) non-vented manual closed chambers. The chambers were pushed ca 3 cm into the soil immediately before the measurement. Each chamber had a rubber septum for gas sampling inserted immediately after chamber installation. Headspace gas samples were collected at the time of chamber deployment and after 60 min. Preliminary investigations on the site showed that the increases in gas concentration within the closed chambers were linear during chamber deployment. Nine gas samples were taken at each section, stored in pre-evacuated 3.5-ml glass vials, transported to the laboratory and analysed immediately. The amount of CH4 was quantified using an HP 5890 Series II gas chromatograph (Hewlett Packard, Palo Alto, CA, USA) equipped with a 2 m Porapak N column at 750°C, and a flame ionization detector. For instrument calibrations standard mixtures of CH4 were used, and the results were corrected for gas losses in connection with transfer to and from glass vials.
Parallel to the gas sampling, true replicate soil samples were obtained from the area of four chambers (0–20 cm depth), by pooling seven subsamples, which were randomly taken using a soil auger of 3-cm diameter. The soil was immediately homogenized by sieving through a 5-mm mesh sieve. Samples for nucleic acid and phospholipid analyses were immediately stored at −80°C and 4°C, respectively.
A higher number of replicate was used for the gas measurements due to the great heterogeneity observed for those areas in preview analysis.
Soil analyses
Gravimetric moisture content was determined after drying at 105°C. Soil pH was measured using a glass electrode in 1:2.5 (w/w) soil/water suspension. Soil mineral N (NH4+, NO3−) was measured colourimetrically from 1 M KCl extracts using a soil (fresh field-moist) solution ratio of 40 g:200 ml (Zbíral et al., 1997). Total organic carbon was determined by wet oxidation with acid dichromate (Jackson, 1958), and total nitrogen content by Kjeldahl digestion (Zbíral, 1995).
Determination of total and archaeal biomass by PLFA and PLEL
Lipids were extracted from fresh soil samples equivalent to 10 g dry weight (d.w.), according to the Bligh–Dyer method as described elsewhere (Zelles and Bai, 1993). The resulting lipid material was fractionated into neutral lipids, glycolipids and phospholipids on a silica-bonded phase column (SPE-SI; Bond Elute, Analytical Chem International, USA) by elution with chloroform, acetone and methanol, respectively. An aliquot of the phospholipid fraction equivalent to 2.5 g d.w. was taken for phospholipid fatty acid (PLFA) analysis. After mild alkaline hydrolysis, described in detail by Oravecz et al. (2004), the resulting fatty acid methyl esters were separated on a capillary gas chromatograph and identified using the MIS automatic identification system (Agilent 6850, FID, TSBA50, MIDI Inc., Newark, DE, USA).
Another aliquot of the phospholipid fraction equivalent to 7.5 g soil d.w. was used for PLEL analysis according to Gattinger et al. (2003). After the formation of ether core lipids, ether-linked isoprenoids were released following cleavage of ether bonds with HI and reductive dehalogenation with Zn in glacial acetic acid. The resulting isoprenoid hydrocarbons were dissolved in 100 μl internal standard solution (nonadecanoic methyl ester) and subjected to gas chromatography-mass spectrometry analysis at operating conditions described elsewhere (Gattinger et al., 2003).
Data of the individual PLFA/PLEL compounds (expressed in nmol g−1 soil d.w.) were added to obtain the concentration of total phospholipid chains, a measure for total microbial biomass in soil (Zelles, 1999). The proportion of individual PLFA/PLEL compounds to the concentration of total phospholipid chains is expressed in % and gives an estimate of the relative abundance of certain microbial groups (Zelles, 1999).
The following abbreviations were used for the different PLEL-derived isoprenoid hydrocarbons: i20:0 indicates a saturated and i20:1 a monounsaturated isoprenoid chain with 20 C atoms; i40:0 denotes an isoprenoid chain with 40 C atoms.
DNA extraction from soil
DNA was extracted from 500 mg soil as described by Griffiths et al. (2000). The quality and quantity of the extracts was evaluated using a spectrophotometer (Nanodrop, PeqLab, Erlangen, Germany).
Real-time quantitative PCR of mcrA genes
The real-time quantitative PCR (qPCR) was performed in an ABI Prism 7700 sequence detection system (Perkin Elmer, Foster City, CA, USA). Reaction mixtures contained 5 μl of qPCR ROX & Go Green (qBiogene, Illkirch, France), 1.5 μg bovine serum albumin (Sigma-Aldrich, Germany), 5 pmol of each primer (Luton et al., 2002), 5% dimethyl sulphoxide (Sigma-Aldrich, Steinheim, Germany), 0.5 μl DNA template and water to a final volume of 25 μl. The amplification was carried out as follows: initial denaturation at 95°C for 15 min and 40 cycles at 94°C for 1 min, 52°C for 1 min and 72°C for 1 min. Standard curves were constructed using 10-fold serial dilutions of plasmids containing a partial sequence of Methanosarcina mazei mcrA gene. Amplification efficiencies of the PCRs were calculated using data from the standard curves with the following formula: efficiency [10(−1/slope)]−1. To discard inhibitory effects of substances co-extracted with the DNA, amplification of serial dilutions were performed for samples of each plot. Four independent assays were run per sample. The quality of the amplification was evaluated by the generation of melting curves of the PCR products and confirmed in 1.5% agarose gels stained with ethidium bromide.
Cloning and sequencing of mcrA genes
Fifty nanograms of DNA were pooled from four replicate extracts of section SI and used as template for the amplification of mcrA genes. The reaction mixture and amplification cycle were performed as described above, excepting the polymerase (Taq polymerase, Invitrogen, Karlsruhe, Germany).
Purified PCR products from the four replicates were pooled and cloned using the TA cloning kit (Invitrogen), following manufacturer's instructions. The reaction contained ca 50 ng of PCR product, 1 μl of ligation buffer, 2.5 μl of sterile dH2O, 2 μl of the vector (pCR 2.1) and 1 μl of T4 ligase. White colonies, containing the insert, were inoculated in Luria–Bertani medium with 50 μl ml−1 kanamycin, grown overnight and used for plasmid isolation (Qiagen Plasmid mini kit, Qiagen, Hilden, Germany). Plasmids holding inserts of correct size were selected after digestion with EcoRI (MBI Fermentas, Vilnius, Lithuania).
Fifty-four clones were randomly selected and sequenced in a ABI Prism 3730 DNA sequencer using the BigDye terminator chemistry (Applied Biosystems, Foster City, CA, USA) according to manufacturer's instructions. Deduced amino-acid sequences of MCR α-subunit (141 positions) were obtained using the program expase and aligned by clustalW. Distance matrix was calculated using ‘Protdist’ (PHYLIP: http://evolution.genetics.washington.edu/phylip.html) and Dayhoff PAM 001 matrix as amino-acid replacement model. Phylogenetic dendrograms deduced MCR sequences from reconstructed distance matrix using neighbour-joining analysis and randomization of species input. Bootstrap values were calculated of 1000 sets.
To estimate the completeness of sampling of a community, rarefaction curves were generated using the software Analytic Rarefaction (http://www.uga.edu/~strata/software/Software.html). Deduced amino-acid sequences showing more than 98% similarity were considered as one operational taxonomic unit (OTU) (Juottonen et al., 2006). The obtained sequences were submitted to National Center for Biotechnology Information database and can be found under the following assession number: DQ994833–DQ994886.
Statistical analysis
All statistical analyses were performed with SPSS software package release 12.0 (SPSS Inc., Chicago, IL, USA). Analysis of variance (ANOVA) was used to determine the significance between treatments followed by all pairwise multiple comparison procedure (Student–Newman–Keuls test). Kruskal–Wallis one-way ANOVA was applied for cases where normality test of data distribution failed.
Results
General soil characteristics and CH4 fluxes
Table 1 compiles the soil properties of the three sections studied. All variables showed significant differences among sections. The increasing levels of organic C (Corg), total N (Nt), ammonium (NH4+) and pH along the gradient from section NI over MI to SI clearly reflected the impact of cattle due to excretal returns. Site SI also showed much higher concentrations of total phospholipids compared to the other sites. The observed differences among the three sections were more pronounced at spring (after cattle left the overwintering area) than at fall sampling (before cattle entered the area).
Table 2 compiles the in situ measurements of CH4 emissions. At both samplings, CH4 emissions at the three sections reflected the different degrees of animal impact on the pasture, that is the highest values in SI and the lowest in NI. Section NI appeared even as a sink for methane. However, the differences among the sites were only statistically significant at spring sampling. CH4 emissions measured for sections SI and MI in fall were much lower than in spring.
Archaeal biomass
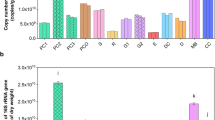
The concentration of archaeal PLEL decreased in accordance with degree of animal impact from section SI to NI (Figure 1a). In spring, PLEL concentration decreased from 28 to 0.5 nmol PLEL isoprenoids g−1 soil d.w., in fall from 28 to 0.7 nmol g−1 soil d.w. The same was true for the relative archaeal abundance (% values), for which the highest values were observed in SI (5.3% and 4.7%) and the lowest in NI (0.4% and 0.7%). Absolute and relative archaeal abundance in section SI remained rather constant, whereas a significant decrease was observed in section MI from spring to fall sampling. For all samples, i20:0 was the most abundant etherlipid, which is a general marker for organism of the phylum Euryarchaeota (Gattinger, 2001). i20:1 isoprenoid chains, detected previously in acetoclastic methanogens from the genus Methanosarcina (Gattinger et al., 2002), were detected in significant amounts in dung (data not shown), SI samples (spring and fall) and in MI samples (only spring).
(a) PLEL composition in soil samples along a gradient of animal impact. (b) Numbers of mcrA gene copies along the same gradient of animal impact of the study site Borova. Bars denote s.d. Values for MI and NI from fall sampling were below detection limit. SI, severe impact; MI, moderate impact; NI, no impact. Per cent values indicate the percentage of PLEL in relation to the concentration of total phospholipid chains. The following abbreviations for the different PLEL-derived isoprenoid hydrocarbons were used: i=isoprenoid branched; 20 and 40=size of the hydrocarbon chain; i20:0 and i20:1=saturated and unsaturated hydrocarbon chains, respectively. Different letters indicate significance of absolute PLEL concentration at the P=0.05 level. PLEL, phospholipid etherlipid.
Abundance and diversity of methanogens
The quantification of mcrA genes revealed a similar pattern as archaeal PLEL concentrations (Figure 1b). In plot SI, the number of target molecules per gram of d.w. soil ranged between 3.8 and 4.2 × 106 copies and was significantly higher than that observed in the other sites at both sampling dates. For plots MI and NI, 4.9 and 2.3 × 105 molecules per gram d.w. soil were detected in spring, respectively. In fall, the values decreased and were below detection limit of approximately 1000 target molecules per gram of d.w. soil. Subsequent qPCR with stepwise dilution of soil DNA revealed no inhibitory substances for PCR in the original nucleic acid extracts (data not shown).
Following the qPCR and PLEL results, only mcrA gene fragments from section SI, with distinct methanogenic characteristics, were subjected to cloning and sequencing. Fifty-four clones were randomly selected for the analysis. The rarefaction's analysis showed that number of clones was sufficient to cover the diversity of mcrA OTUs. Phylogenetic analysis of mcrA clones revealed the presence of two main clusters (Figure 2a). Cluster II comprises sequences affiliated with cultured methanogens. The majority of the sequences within this cluster show close similarities to Methanothermus, Methanosaeta and Methanosarcina and represents 15%, 11% and 33% of the analysed sequences, respectively (Figure 2b). Cluster I contains sequences only from uncultured rumen methanogens. Twenty-two sequences from soil section SI (41% of the total, Figure 2b) and three sequences from uncultured rumen archaeons are forming this cluster.
(a) Dendrogram showing phylogenetic relationships of mcrA sequences of soil samples collected in spring at the severe animal impacted (SI) area. The tree was constructed from inferred amino-acid sequences (141 positions) using Dayhoff PAM 001 distance matrix and neighbour-joining analysis. Numbers at the nodes represent the percentages of bootstrap resamplings based on 1000 replicates. Methanopyrus kandleri was used as outgroup reference. (b) Distribution of partial sequences from mcrA genes retrieved from SI.
Discussion
Upland land soils, including grassland soils, are normally described as sink for the green house gas CH4 (Hütsch et al., 1994; Castaldi et al., 2007). Nevertheless, as shown in the present study, keeping cattle in overwintering pastures may cause enormous changes in soil properties, mainly due to excretal returns and treading (Oenema et al., 2001; White et al., 2001), which in turn stimulate CH4 production.
In spring, after the animals have been removed from the pasture, microbial biomass, organic C and total N values were higher in section SI followed by MI and NI, showing a clear relationship between the degree of impact and changes in soil properties and soil microbial community. Although, in fall (5 months after the animals have been removed) biomass values remained high in section SI, a clear reduction was observed in MI. Hernández et al. (2007) and collaborators postulated that animal excrements stimulate growth of soil microorganism, however high inputs are necessary to maintain an elevated microbial biomass content.
Archaeal biomass measurements showed the same trend as observed for total microbial biomass. In section NI, the concentration of PLEL was 0.6 in spring and 0.8 nmol g−1 dry matter in fall and contributed to 0.4% and 0.7% of total phospholipid chains of that section. These values are in the range of previously reported data from light or unfertilized arable soils (Gattinger et al., 2002). In section SI, values were well above the range determined so far for any upland (Gattinger et al., 2007) or wetland soil (Bai et al., 2000; Wachinger et al., 2000). Wachinger et al. (2000) suggested that archaeal abundance reflects in situ and in vitro CH4 production/emission potential, even though a linear correlation could not be established. This result is not surprising as archaeal biomass is probably not an ideal indicator for methanogenesis, as only a fraction of the whole community is able to carry out this process.
The determination of the mcrA gene gives further insights into the functional role of the detected archaea. As until now, no quantitative data on mcrA gene copies have been reported for a similar terrestrial environment, comparisons in this respect can hardly be made. However, it must be taken into account that the measured values might be biased by the used primers and the nucleic acid extraction efficiency (Sharma et al., 2007), although several protocols were tested using soil samples from the corresponding farm, indicating that the chosen protocol for this study gave the highest and most reproducible values. Therefore, it was of great importance to have quantitative data from a second, PCR independent marker (etherlipids), which show similar trends compared to the molecular data. Overall, the number of mcrA gene copies showed the same trend as has been described for the archaeal phospholipids: highest values in section SI and lowest in NI. Also the decrease in PLEL concentration in MI from spring to fall sampling went parallel with the decrease in the number of mcrA gene copies from spring to fall in MI. Moreover, although qPCR can provide accurate measurements of gene numbers per unit of soil, it is more difficult to convert these values into cell densities and biomass or to set them in direct relation to soil mass or soil volume. A conversion to cell densities requires the knowledge of the copy number per genome and the genome size (Sharma et al., 2007).
Quantification of mcrA genes may be better indicator for methanogenesis than the total archaea biomass; nevertheless, it only represents the potential of the community, as it is based on the DNA level. This assertion can be confirmed by the analysis of fall data for section SI, in which methanogenic biomass values remained high even though a harsh decrease in methane emissions was observed.
Phylogenetic analysis revealed the presence of a diverse methanogenic community in section SI. Most of the detected methanogenic taxa were also found in arable soils fertilized with cattle manure for more than 50 years (Gattinger et al., 2007). Approximately 41% of the sequences formed a separate cluster and showed close relationships (97% based on DNA sequences) to uncultured rumen archaeons (Tatsuoka et al., 2004). Moreover, 33% were similar to Methanosarcinaceae representatives, which are known to occur in cattle rumen (Jarvis et al., 2000). Hence, a substantial part of the retrieved clones from section SI can be considered as typical rumen colonizers. Lipids normally present in the membrane of Methanosarcina (i20:1 side chain; Gattinger et al., 2002) were also detected in significant amounts in fresh excrements sampled from the overwintering area (data not shown). PLEL profiling revealed at both samplings the highest concentration of Methanosarcinaceae-derived lipids in section SI. These findings support the hypothesis that with the cattle excrements ‘rumenborne’ methanogens are transferred to the grassland soil. On the other hand, Methanosarcinaceae are generally considered as somewhat ubiquitous methanogens occurring in oxic and anoxic soils of the temperate zone (Lueders et al., 2001; Ramakrishnan et al., 2001). Signature lipids of this group have also been detected, although in low amounts, in the non-impacted section NI.
The combination of the obtained data from physicochemical, methane emission, phospholipid and nucleic acid analyses gives strong indication of the following phenomenon: during the overwintering period, cattle stay most of the time in section SI, where they excrete faeces and urine. As a result of the urine input, the soil pH rises from slightly acidic to alkaline. A pH of 8 is known to be optimal for the activity of acetoclastic methanogens like Methanosarcinaceae (Beeman and Suflita, 1990). Supplied with easily available organic substrates from the cow dung, soilborne methanogens like Methanosarcina spp are stimulated in growth and activity. In addition, hitherto unknown, ‘rumenborne’ archaea along with representatives from known methanogenic groups including Methanosarcina species are transferred from the cattle intestinal tract to the soil. In this terrestrial habitat, these microorganisms will find suitable conditions for growth and methanogenic activity. A similar phenomenon was also described for arable soils receiving high amounts of cattle manure (Gattinger et al., 2007). However, in this case, methane emissions were by far less pronounced, presumably because of significant methanotrophic activities. The reasons for the low activity of methane oxidizers are not clear. On the one hand, it is known that high concentration of ammonia, originated in our experiment from cattle urine, could inhibit methane oxidation (Hütsch et al., 1993; Schnell and King, 1994). On the other hand, it could be argued that the ratio between methane in the soil and ammonium is so high that the methane oxidizers are not inhibited by the high ammonium concentrations but even suffer from nitrogen limitation, which reduces their activity (Chan and Parkin, 2001). Moreover, in section SI, an increase in anaerobic micro-sites cased by animal treading and excrements, could also affect this microbial group. In any case, methane oxidation by low-affinity methane oxidizers or anaerobic methane oxidation may play an important role on site SI. However, this aspect is not addressed in the present study.
For the moderate impacted section MI, we assumed that the same phenomenon occurred, but to a far lesser extent. Only at spring, after the cattle left the overwintering area, a significant methanogenic biomass was observed, as indicated by the high abundance of mcrA genes. This presumably occurred because of suitable soil ecological properties (pH and organic C), leading to a distinct methane production. Nevertheless, after 6 months of no cattle impact, methanogenic biomass declined sharply and mcrA genes were not detectable along with a clear reduction in pH and organic C content, which were almost the same as in section NI. Owing to these changes in soil characteristics, a far lesser methane emission in section MI was observed at fall than at spring sampling.
In contrast, in SI, the methanogenic community sustained its biomass, as indicated by the abundance of the mcrA genes, during a 6 month cattle-free period. Also organic C and pH remained at a high level providing an appropriate habitat for methanogenic archaea. Nevertheless, as postulated by Plaza (Plaza et al., 2007) and collaborators, high amounts of easily degradable organic matter are introduced in soil via animal excrement, which in turn increases microbial activity. Once this nutrient source is no longer available, the community adapts to the new environmental condition and reduces its metabolic activity. Moreover, the abundance of phospholipid biomarkers for anaerobic bacteria was higher in spring compared to fall, indicating changes in overall oxygen availability probably due to a recovery of the highly compacted soils in spring. However, SI plots showed strong aggregate formation during summer (data not shown). This is not surprising as soils of high pH and high carbon concentrations are known to aggregate easily (Boix-Fayos et al., 2001). Also the influence of cattle manure on soil aggregate formation is well known (Pare et al., 1999). Not only the high content of carbon itself but also the resulting increases in microbial biomass and activity may stimulate soil aggregation (Martens and Frankenberger, 1992). The aggregate interior, with low oxygen concentrations may be a good habitat for methanogenic microbes to survive the unfavourable summer conditions. In MI, the recovery of vegetation during the cattle-free period was very much pronounced. Obviously, the methanogenic Archaea are not highly adapted to grassland rhizosphere and were outcompeted by other microbes. To clarify more the relation between microbial activity and methane production, mRNA-based studies indicating the induction of gene expression of mcrA are necessary. The results of this study are based on DNA studies which are reflecting more a genetic potential than an actual activity.
It can be concluded that after a period of at least 6 months of no cattle impact, the formerly, severely impacted section maintained its methane production potential, whereas the methane production potential under moderate impact returned to background values.
References
Bai Q, Gattinger A, Zelles L . (2000). Characterisation of microbial consortia in paddy rice soil by phosholipid analysis. Microb Ecol 39: 273–281.
Beeman RE, Suflita JM . (1990). Environmental factors influencing methanogenesis in a shallow anoxic aquifer: a field and laboratory study. J Ind Microbiol 5: 45–57.
Boix-Fayos C, Calvo-Cases A, Imeson AC . (2001). Influence of soil properties on the aggregation of some Mediterranean soils and the use of aggregate size and stability as land degradation indicators. Catene 44: 47–67.
Castaldi S, Costantini M, Cenciarelli P, Ciccioli P, Valentini R . (2007). The methane sink associated to soils of natural and agricultural ecosystems in Italy. Chemosphere 66: 723–729.
Chan ASK, Parkin TB . (2001). Methane oxidation and production activity in soils from natural and agricultrual ecosystems. J Environ Qual 301: 1896–1903.
Clayton H, Arah JRM, Smith KA . (1994). Measurement of nitrous oxide emissions from fertilized grassland using closed chambers. J Geophys Res 99: 599–607.
Ellermann J, Hedderich R, Bocher R, Thauer RK . (1988). The final step in methane formation. Investigations with highly purified methyl-CoM reductase (component C) from Methanobacterium thermoautotrophicum (strain Marburg). Eur J Biochem 172: 669–677.
Gattinger A . (2001). Entwicklung und Anwendung von Methoden zur Charakterisierung von mikrobiellen Gemeischaften in oxischen und Bodenökosystem anhand von Phospholipid-Profilen. Fakultät Wissenschaftszentrum Weihehstephan für Ernährung, Landnutzung und Umwelt. Technische Universität München: Munich, p 147.
Gattinger A, Hofle MG, Schloter M, Embacher A, Bohme F, Munch JC et al. (2007). Traditional cattle manure application determines abundance, diversity and activity of methanogenic Archaea in arable European soil. Environ Microbiol 9: 612–624.
Gattinger A, Günthner A, Schloter M, Munch JC . (2003). Characterization of Archaea in soils by polar lipid analysis. Acta Biotechnol 23: 21–28.
Gattinger A, Ruser R, Schloter M, Munch JC . (2002). Microbial community structure varies in different soil zones of a potato field. J Plant Nutr Soil Sci 165: 421–428.
Griffiths RI, Whiteley AS, O’Donnell AG, Bailey MJ . (2000). Rapid method for coextraction of DNA and RNA from natural environments for analysis of ribosomal DNA- and rRNA-based microbial community composition. Appl Environ Microbiol 66: 5488–5491.
Hernández D, Fernández JM, Plaza C, Polo A . (2007). Water-soluble organic matter and biological activity of a degraded soil amended with pig slurry. Sci Total Environ (in press).
Hütsch BW, Webster CP, Powlson DS . (1993). Long-term effects of nitrogen fertilization on methane oxidation in soil of the broadbalk wheat experiment. Soil Biol Biochem 25: 1307–1310.
Hütsch BW, Webster CP, Powlson DS . (1994). Methane oxidation in soil as affected by land use, soil pH and N fertilization. Soil Biol Biochem 26: 1613–1622.
Jackson ML . (1958). Soil Chemical Analysis. Prentice Hall: Englewood Cliffs: New York.
Jarvis GN, Strompl C, Burgess DM, Skillman LC, Moore ER, Joblin KN . (2000). Isolation and identification of ruminal methanogens from grazing cattle. Curr Microbiol 40: 327–332.
Juottonen H, Galand PE, Yrjälä K . (2006). Detection of methanogenic Archaea in peat: comparison of PCR primers targeting the mcrA gene. Res Microbiol 15: 914–921.
Lueders T, Chin K-J, Conrad R, Friedrich M . (2001). Molecular analyses of methyl-coenzyme M reductase α-subunit (mcrA) genes in rice field soil and enrichment cultures reveal the methanogenic phenotype of a novel archaeal lineage. Environ Microbiol 3: 194–204.
Luton PE, Wayne JM, Sharp RJ, Riley PW . (2002). The mcrA gene as an alternative to 16S rRNA in the phylogenetic analysis of methanogen populations in ladfill. Microbiology 148: 3521–3530.
Martens DA, Frankenberger WT . (1992). Modification of infiltration rates in an organic-amended irrigated soil. Agron J 84: 707–717.
Menneer JC, Ledgard S, McLay C, Silverster W . (2005). Animal treading stimulates denitrification in soil under pasture. Soil Biol Biochem 37: 1625–1629.
Mosier A, Schimel D, Valentine D, Bronson K, Parton W . (1991). Methane and nitrous oxide fluxes in native, fertilized and cultivated grasslands. Nature 350: 330–332.
OCA (2006). Organic agriculture can help stabilize global climate change. Organic Consumers Association, http://www.organicconsumers.org/organic/stabalize062404.cfm.
Oenema O, Velthof G, Kuikman P . (2001). Technical and policy aspects of strategies to decrease greenhouse gas emissions from agriculture. Nutr Cycl Agroecosys 60: 301–315.
Oravecz O, Elhottovás D, Kristufek V, Sustr V, Frouz J, Triska J et al. (2004). Application of ARDRA and PLFA analysis in characterizing the bacterial communities of the food, gut and excrement of saprophagous larvae of Penthetria holosericea (Diptera: Bibionidae): a pilot study. Folia Microbiol (Praha) 49: 83–93.
Pare T, Dinel H, Moulin AP, Townley-Smith L . (1999). Organic matter quality and structural stability of a Black Chernozemic soil under different manure and tillage practices. Geoderma 91: 311–326.
Plaza C, García-Gil JC, Polo A . (2007). Microbial activity in pig slurry-amended soils under aerobic incubation. Biodegradation 18: 159–165.
Ramakrishnan B, Lueders T, Dunfield PF, Conrad R, Friedrich MW . (2001). Archaeal community structures in rice soils from different geographical regions before and after initiation of methane production. FEMS Microbiol Ecol 37: 175–186.
Schnell S, King GM . (1994). Mechanistic analysis of ammonium inhibition of atmospheric methane consumption in forest soils. Appl Environ Microbiol 60: 3514–3521.
Sharma S, Radl V, Hai B, Kloos K, Fuka M, Engel M et al. (2007). Quantification of functional genes from procaryotes in soil by PCR. J Microbiol Methods 68: 445–452.
Sherlock RR, Sommer SG, Khan RZ, Wood CW, Guertal EA, Freney JR et al. (2002). Ammonia, methane, and nitrous oxide emission from pig slurry applied to a pasture in New Zealand. J Environ Qual 31: 1491–1501.
Šimek M, Brucek P, Hynšt J, Uhlírová E, Petersen SO. (2006). Effects of excretal returns and soil compaction on nitrous oxide emissions from a cattle overwintering area. Agr Ecosys Environ 112: 186–191.
Sommer SG, Sherlock RR, Khan RZ . (1996). Nitrous oxide and methane emissions from pig slurry amended soils. Soil Biol Biochem 28: 1541–1544.
Tatsuoka N, Mohammed N, Mitsumori M, Hara K, Kurihara M, Itabashi H . (2004). Phylogenetic analysis of methyl coenzyme-M reductase detected from the bovine rumen doi:10.1111/j.1472-765X.2004.01566.x. Lett Appl Microbiol 39: 257–260.
Thauer R . (1998). Biochemistry of methanogenesis: a tribute to Marjory Stephenson. 1998 Marjory Stephenson Prize Lecture. Microbiology 144: 2377–2406.
Wachinger G, Fiedler S, Zepp K, Gattinger A, Sommer M, Roth K . (2000). High spatial variability of methane production on the micro-scale: spatial association with hot spots of organic material and archaeal populations. Soil Biol Biochem 32: 1121–1130.
Wagner D, Lipski A, Embacher A, Gattinger A . (2005). Methane fluxes in permafrost habitats of the Lena Delta: effects of microbial community structure and organic matter quality. Environ Microbiol 7: 1582–1592.
White SL, Sheffield RE, Washburn SP, King LD, Green Jr JT . (2001). Spatial and time distribution of dairy cattle excreta in an intensive pasture system. J Environ Qual 30: 2180–2187.
Zbíral J . (1995). Soil Analyses, Part I. Czech Central Institute for Supervising and Testing in Agriculture: Brno (in Czech).
Zbíral J, Honsa I, Malý S . (1997). Soil Analyses, Part III. Czech Central Institute for Supervising and Testing in Agriculture: Brno (in Czech).
Zelles L . (1999). Identification of single cultured micro-organisms based on their whole-community fatty acid profiles, using an extended extraction procedure. Chemosphere 39: 665–682.
Zelles L, Bai QY . (1993). Fractionation of fatty acids derived from soil lipids by soil phase extraction and their quantitative analysis by GC-MS. Soil Biol Biochem 25: 130–134.
Acknowledgements
This study was supported by a grant from the DAAD (German academic exchange program) and by the Ministry of Education of the Czech Republic (LC06066) Grant Agency of the Academy of Sciences of the Czech Republic (IAA600660605) the Research Plan of the Biology Centre, Institute of Soil Biology (AV 0Z 60660521). V Šlajchrtová, E Zadáková and Conny Galonska are greatly acknowledged for field work and laboratory analyses. Mr and Mrs Kamír are thanked for allowing access to the experimental plot and for their significant support in field work. We also wish to thank the two anonymous reviewers for their helpful comments.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Radl, V., Gattinger, A., Chroňáková, A. et al. Effects of cattle husbandry on abundance and activity of methanogenic archaea in upland soils. ISME J 1, 443–452 (2007). https://doi.org/10.1038/ismej.2007.60
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ismej.2007.60
Keywords
This article is cited by
-
The impact of long-term organic farming on soil-derived greenhouse gas emissions
Scientific Reports (2019)
-
Soil carbon dioxide and methane fluxes from forests and other land use types in an African tropical montane region
Biogeochemistry (2019)
-
Dung application increases CH4 production potential and alters the composition and abundance of methanogen community in restored peatland soils from Europe
Biology and Fertility of Soils (2018)
-
Evaluating the lingering effect of livestock grazing on functional potentials of microbial communities in Tibetan grassland soils
Plant and Soil (2016)
-
Manure-associated stimulation of soil-borne methanogenic activity in agricultural soils
Biology and Fertility of Soils (2015)