Abstract
Background/Objectives:
There is increasing evidence of a relationship between blood DNA methylation and body mass index (BMI). We aimed to assess associations of BMI with individual methylation measures (CpGs) through a cross-sectional genome-wide DNA methylation association study and a longitudinal analysis of repeated measurements over time.
Subjects/Methods:
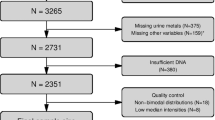
Using the Illumina Infinium HumanMethylation450 BeadChip, DNA methylation measures were determined in baseline peripheral blood samples from 5361 adults recruited to the Melbourne Collaborative Cohort Study (MCCS) and selected for nested case–control studies, 2586 because they were subsequently diagnosed with cancer (cases) and 2775 as controls. For a subset of 1088 controls, these measures were repeated using blood samples collected at wave 2 follow-up, a median of 11 years later; weight was measured at both time points. Associations between BMI and blood DNA methylation were assessed using linear mixed-effects regression models adjusted for batch effects and potential confounders. These were applied to cases and controls separately, with results combined through fixed-effects meta-analysis.
Results:
Cross-sectional analysis identified 310 CpGs associated with BMI with P<1.0 × 10−7, 225 of which had not been reported previously. Of these 225 novel associations, 172 were replicated (P<0.05) using the Atherosclerosis Risk in Communities (ARIC) study. We also replicated using MCCS data (P<0.05) 335 of 392 associations previously reported with P<1.0 × 10−7, including 60 that had not been replicated before. Associations between change in BMI and change in methylation were observed for 34 of the 310 strongest signals in our cross-sectional analysis, including 7 that had not been replicated using the ARIC study.
Conclusions:
Together, these findings suggest that BMI is associated with blood DNA methylation at a large number of CpGs across the genome, several of which are located in or near genes involved in ATP-binding cassette transportation, tumour necrosis factor signalling, insulin resistance and lipid metabolism.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout

Similar content being viewed by others
References
Meissner A, Mikkelsen TS, Gu H, Wernig M, Hanna J, Sivachenko A et al. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature 2008; 454: 766–770.
van Dongen J, Nivard MG, Willemsen G, Hottenga JJ, Helmer Q, Dolan CV et al. Genetic and environmental influences interact with age and sex in shaping the human methylome. Nat Commun 2016; 7: 11115.
Fraga MF, Esteller M . Epigenetics and aging: the targets and the marks. Trends Genet 2007; 23: 413–418.
Boks MP, Derks EM, Weisenberger DJ, Strengman E, Janson E, Sommer IE et al. The relationship of DNA methylation with age, gender and genotype in twins and healthy controls. PLoS One 2009; 4: e6767.
Zhang FF, Cardarelli R, Carroll J, Fulda KG, Kaur M, Gonzalez K et al. Significant differences in global genomic DNA methylation by gender and race/ethnicity in peripheral blood. Epigenetics 2011; 6: 623–629.
Shenker NS, Polidoro S, van Veldhoven K, Sacerdote C, Ricceri F, Birrell MA et al. Epigenome-wide association study in the European Prospective Investigation into Cancer and Nutrition (EPIC-Turin) identifies novel genetic loci associated with smoking. Hum Mol Genet 2013; 22: 843–851.
Zeilinger S, Kuhnel B, Klopp N, Baurecht H, Kleinschmidt A, Gieger C et al. Tobacco smoking leads to extensive genome-wide changes in DNA methylation. PLoS ONE 2013; 8: e63812.
Guida F, Sandanger TM, Castagne R, Campanella G, Polidoro S, Palli D et al. Dynamics of smoking-induced genome-wide methylation changes with time since smoking cessation. Hum Mol Genet 2015; 24: 2349–2359.
Robertson KD . DNA methylation and human disease. Nat Rev Genet 2005; 6: 597–610.
Esteller M . Epigenetics in cancer. N Engl J Med 2008; 358: 1148–1159.
Chambers JC, Loh M, Lehne B, Drong A, Kriebel J, Motta V et al. Epigenome-wide association of DNA methylation markers in peripheral blood from Indian Asians and Europeans with incident type 2 diabetes: a nested case–control study. Lancet Diabetes Endocrinol 2015; 3: 526–534.
Guenard F, Tchernof A, Deshaies Y, Cianflone K, Kral JG, Marceau P et al. Methylation and expression of immune and inflammatory genes in the offspring of bariatric bypass surgery patients. J Obes 2013; 2013: 492170.
Pigeyre M, Yazdi FT, Kaur Y, Meyre D . Recent progress in genetics, epigenetics and metagenomics unveils the pathophysiology of human obesity. Clin Sci (London, England: 1979) 2016; 130: 943–986.
Sharp GC, Lawlor DA, Richmond RC, Fraser A, Simpkin A, Suderman M et al. Maternal pre-pregnancy BMI and gestational weight gain, offspring DNA methylation and later offspring adiposity: findings from the Avon Longitudinal Study of Parents and Children. Int J Epidemiol 2015; 44: 1288–1304.
van Dijk SJ, Molloy PL, Varinli H, Morrison JL, Muhlhausler BS . Epigenetics and human obesity. Intern J Obes 2015; 39: 85–97.
Dick KJ, Nelson CP, Tsaprouni L, Sandling JK, Aissi D, Wahl S et al. DNA methylation and body-mass index: a genome-wide analysis. Lancet (London, England) 2014; 383: 1990–1998.
Demerath EW, Guan W, Grove ML, Aslibekyan S, Mendelson M, Zhou YH et al. Epigenome-wide association study (EWAS) of BMI, BMI change and waist circumference in African American adults identifies multiple replicated loci. Hum Mol Genet 2015; 24: 4464–4479.
Aslibekyan S, Demerath EW, Mendelson M, Zhi D, Guan W, Liang L et al. Epigenome-wide study identifies novel methylation loci associated with body mass index and waist circumference. Obesity (Silver Spring, Md.) 2015; 23: 1493–1501.
Wahl S, Drong A, Lehne B, Loh M, Scott WR, Kunze S et al. Epigenome-wide association study of body mass index, and the adverse outcomes of adiposity. Nature 2017; 541: 81–86.
Mendelson MM, Marioni RE, Joehanes R, Liu C, Hedman AK, Aslibekyan S et al. Association of body mass index with DNA methylation and gene expression in blood cells and relations to cardiometabolic disease: a Mendelian randomization approach. PLoS Med 2017; 14: e1002215.
Ali O, Cerjak D, Kent Jr JW, James R, Blangero J, Carless MA et al. Methylation of SOCS3 is inversely associated with metabolic syndrome in an epigenome-wide association study of obesity. Epigenetics 2016; 1–9.
Al Muftah WA, Al-Shafai M, Zaghlool SB, Visconti A, Tsai PC, Kumar P et al. Epigenetic associations of type 2 diabetes and BMI in an Arab population. Clin Epigenet 2016; 8: 13.
Wilson LE, Harlid S, Xu Z, Sandler DP, Taylor JA . An epigenome-wide study of body mass index and DNA methylation in blood using participants from the Sister Study cohort. Int J Obes (2005) 2017; 41: 194–199.
Milne RL, Fletcher AS, MacInnis RJ et al. Cohort Profile: the Melbourne Collaborative Cohort Study (Health 2020). Int J Epidemiol 2017 doi:10.1093/ije/dyx085. [Epub ahead of print 2017 June 21].
Lohman TG, Roche AF, Martorell R . Anthropometric Standardization Reference Manual. Human Kinetics Books: Champaign, IL, 1988.
Bassett JK, Baglietto L, Hodge AM, Severi G, Hopper JL, English DR et al. Dietary intake of B vitamins and methionine and breast cancer risk. Cancer Causes Control 2013; 24: 1555–1563.
Joo JE, Wong EM, Baglietto L, Jung CH, Tsimiklis H, Park DJ et al. The use of DNA from archival dried blood spots with the Infinium HumanMethylation450 array. BMC Biotechnol 2013; 13: 23.
Aryee MJ, Jaffe AE, Corrada-Bravo H, Ladd-Acosta C, Feinberg AP, Hansen KD et al. Minfi: a flexible and comprehensive Bioconductor package for the analysis of Infinium DNA methylation microarrays. Bioinformatics (Oxford, England) 2014; 30: 1363–1369.
Maksimovic J, Gordon L, Oshlack A . SWAN: Subset-quantile within array normalization for illumina infinium HumanMethylation450 BeadChips. Genome Biol 2012; 13: R44.
Dugue PA, English DR, MacInnis RJ, Jung CH, Bassett JK, FitzGerald LM et al. Reliability of DNA methylation measures from dried blood spots and mononuclear cells using the HumanMethylation450k BeadArray. Sci Rep 2016; 6: 30317.
Houseman EA, Accomando WP, Koestler DC, Christensen BC, Marsit CJ, Nelson HH et al. DNA methylation arrays as surrogate measures of cell mixture distribution. BMC Bioinform 2012; 13: 86.
Viechtbauer W . Conducting meta-analyses in R with the metafor package. J Stat Softw 2010; 36: 03.
Peters TJ, Buckley MJ, Statham AL, Pidsley R, Samaras K, V Lord R et al. De novo identification of differentially methylated regions in the human genome. Epigenet Chromatin 2015; 8: 6.
Johnson WE, Li C, Rabinovic A . Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics (Oxford, England) 2007; 8: 118–127.
Glymour MM, Weuve J, Berkman LF, Kawachi I, Robins JM . When is baseline adjustment useful in analyses of change? An example with education and cognitive change. Am J Epidemiol 2005; 162: 267–278.
Phipson B, Maksimovic J, Oshlack A . missMethyl: an R package for analyzing data from Illumina's HumanMethylation450 platform. Bioinformatics (Oxford, England) 2016; 32: 286–288.
Kanehisa M, Goto S . KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res 2000; 28: 27–30.
Greenland S . Quantifying biases in causal models: classical confounding vs collider-stratification bias. Epidemiology (Cambridge, Mass.) 2003; 14: 300–306.
van Iterson M, van Zwet WE, Consortium B, Heijmans BT . Controlling bias and inflation in epigenome- and transcriptome-wide association studies using the empirical null distribution. Genome Biol 2017; 18: 19.
Huang YT, Chu S, Loucks EB, Lin CL, Eaton CB, Buka SL et al. Epigenome-wide profiling of DNA methylation in paired samples of adipose tissue and blood. Epigenetics 2016; 11: 227–236.
Barfield RT, Almli LM, Kilaru V, Smith AK, Mercer KB, Duncan R et al. Accounting for population stratification in DNA methylation studies. Genet Epidemiol 2014; 38: 231–241.
Hannum G, Guinney J, Zhao L, Zhang L, Hughes G, Sadda S et al. Genome-wide methylation profiles reveal quantitative views of human aging rates. Mol Cell 2013; 49: 359–367.
Gao X, Jia M, Zhang Y, Breitling LP, Brenner H . DNA methylation changes of whole blood cells in response to active smoking exposure in adults: a systematic review of DNA methylation studies. Clin Epigenet 2015; 7: 113.
Issa JP . Aging and epigenetic drift: a vicious cycle. J Clin Invest 2014; 124: 24–29.
Breitling LP, Yang R, Korn B, Burwinkel B, Brenner H . Tobacco-smoking-related differential DNA methylation: 27K discovery and replication. Am J Hum Genet 2011; 88: 450–457.
Pfeiffer L, Wahl S, Pilling LC, Reischl E, Sandling JK, Kunze S et al. DNA methylation of lipid-related genes affects blood lipid levels. Circ Cardiovasc Genet 2015; 8: 334–342.
Kriebel J, Herder C, Rathmann W, Wahl S, Kunze S, Molnos S et al. Association between DNA methylation in whole blood and measures of glucose metabolism: KORA F4 Study. PLoS One 2016; 11: e0152314.
Hidalgo B, Irvin MR, Sha J, Zhi D, Aslibekyan S, Absher D et al. Epigenome-wide association study of fasting measures of glucose, insulin, and HOMA-IR in the Genetics of Lipid Lowering Drugs and Diet Network study. Diabetes 2014; 63: 801–807.
Yvan-Charvet L, Wang N, Tall AR . Role of HDL, ABCA1, and ABCG1 transporters in cholesterol efflux and immune responses. Arterioscler Thromb Vasc Biol 2010; 30: 139–143.
Frisdal E, Le Lay S, Hooton H, Poupel L, Olivier M, Alili R et al. Adipocyte ATP-binding cassette G1 promotes triglyceride storage, fat mass growth, and human obesity. Diabetes 2015; 64: 840–855.
Frisdal E, Le Goff W . Adipose ABCG1: a potential therapeutic target in obesity? Adipocyte 2015; 4: 315–318.
Tzanavari T, Giannogonas P, Karalis KP . TNF-alpha and obesity. Curr Direct Autoimmun 2010; 11: 145–156.
Nieto-Vazquez I, Fernandez-Veledo S, Kramer DK, Vila-Bedmar R, Garcia-Guerra L, Lorenzo M . Insulin resistance associated to obesity: the link TNF-alpha. Arch Physiol Biochem 2008; 114: 183–194.
Yang L, Li P, Fu S, Calay ES, Hotamisligil GS . Defective hepatic autophagy in obesity promotes ER stress and causes insulin resistance. Cell Metab 2010; 11: 467–478.
Acknowledgements
This work was supported by the Australian National Health and Medical Research Council (NHMRC) (grant 1088405). MCCS cohort recruitment was funded by VicHealth and Cancer Council Victoria. The MCCS was further supported by Australian NHMRC grants 209057 and 396414 and by infrastructure provided by Cancer Council Victoria. Cases were ascertained through the Victorian Cancer Registry (VCR) and the Australian Cancer Database (Australian Institute of Health and Welfare). The nested case-control methylation studies were supported by the NHMRC grants 1011618, 1026892, 1027505, 1050198, 1043616 and 1074383. MCS is an NHMRC Senior Research Fellow (1061177). The Atherosclerosis Risk in Communities study has been funded in whole or in part with Federal funds from the National Heart, Lung, and Blood Institute, National Institutes of Health, Department of Health and Human Services (contract numbers HHSN268201700001I, HHSN268201700002I, HHSN268201700003I, HHSN268201700004I and HHSN268201700005I). The authors thank the staff and participants of the ARIC study for their important contributions. Funding support for 'Building on GWAS for NHLBI-diseases: the US CHARGE consortium' was provided by the NIH through the American Recovery and Reinvestment Act of 2009 (ARRA) (5RC2HL102419). Funding support for 'Building on GWAS for NHLBI-diseases: the US CHARGE consortium' was provided by the NIH through the American Recovery and Reinvestment Act of 2009 (ARRA) (5RC2HL102419). YMG was supported by a Radboud University Individual Travel Grant and funds from the Radboudumc Student budget. LB was supported by a Marie Curie International Incoming Fellowship within the 7th European Community Framework Programme. We thank the staff and participants of the MCCS and ARIC studies for their important contributions.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on International Journal of Obesity website
Supplementary information
Rights and permissions
About this article
Cite this article
Geurts, Y., Dugué, PA., Joo, J. et al. Novel associations between blood DNA methylation and body mass index in middle-aged and older adults. Int J Obes 42, 887–896 (2018). https://doi.org/10.1038/ijo.2017.269
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ijo.2017.269
This article is cited by
-
Characterisation of ethnic differences in DNA methylation between UK-resident South Asians and Europeans
Clinical Epigenetics (2022)
-
Methylation-based markers of aging and lifestyle-related factors and risk of breast cancer: a pooled analysis of four prospective studies
Breast Cancer Research (2022)
-
Placental DNA methylation changes associated with maternal prepregnancy BMI and gestational weight gain
International Journal of Obesity (2020)
-
DNA methylation and body mass index from birth to adolescence: meta-analyses of epigenome-wide association studies
Genome Medicine (2020)
-
Epigenome-wide association study in peripheral white blood cells involving insulin resistance
Scientific Reports (2019)