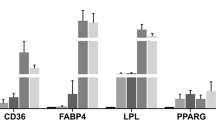
Abstract
Background:
It is well known that high-fat diet (HFD) can cause immune system-related pathological alterations after a significant body weight gain. The mechanisms of the delayed pathological alterations during the development of diet-induced obesity (DIO) are not fully understood.
Methods:
To elucidate the mechanisms underlying DIO development, we analyzed time-course microarray data obtained from a previous study. First, differentially expressed genes (DEGs) were identified at each time point by comparing the hepatic transcriptome of mice fed HFD with that of mice fed normal diet. Next, we clustered the union of DEGs and identified annotations related to each cluster. Finally, we constructed an ‘integrated obesity-associated gene regulatory network (GRN) in murine liver’. We analyzed the epididymal white adipose tissue (eWAT) transcriptome usig the same procedure.
Results:
Based on time-course microarray data, we found that the genes associated with immune responses were upregulated with an oscillating expression pattern between weeks 2 and 8, relatively downregulated between weeks 12 and 16, and eventually upregulated after week 20 in the liver of the mice fed HFD. The genes associated with immune responses were also upregulated at late stage, in the eWAT of the mice fed HFD. These results suggested that a critical transition occurred in the immune system-related transcriptomes of the liver and eWAT around week 16 of the DIO development, and this may be associated with the delayed pathological alterations. The GRN analysis suggested that Maff may be a key transcription factor for the immune system-related critical transition thatoccurred at week 16. We found that transcription factors associated with immune responses were centrally located in the integrated obesity-associated GRN in the liver.
Conclusions:
In this study, systems analysis identified regulatory network modules underlying the delayed immune system-related pathological changes during the development of DIO and could suggest possible therapeutic targets.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Bouchard C . Gene-environment interactions in the etiology of obesity: defining the fundamentals. Obesity 2008; 16: S5–S10.
Phillips CM, Tierney AC, Perez-Martinez P, Defoort C, Blaak EE, Gjelstad IM et al. Obesity and body fat classification in the metabolic syndrome: impact on cardiometabolic risk metabotype. Obesity 2013; 21: E154–E161.
Lin S, Thomas TC, Storlien LH, Huang XF . Development of high fat diet-induced obesity and leptin resistance in C57Bl/6 J mice. Int J Obes Relat Metab Disord 2000; 24: 639–646.
Petro AE, Cotter J, Cooper DA, Peters JC, Surwit SJ, Surwit RS . Fat, carbohydrate, and calories in the development of diabetes and obesity in the C57BL/6 J mouse. Metabolism 2004; 53: 454–457.
Rossmeisl M, Rim JS, Koza RA, Kozak LP . Variation in type 2 diabetes—related traits in mouse strains susceptible to diet-induced obesity. Diabetes 2003; 52: 1958–1966.
Do GM, Oh HY, Kwon EY, Cho YY, Shin SK, Park HJ et al. Long-term adaptation of global transcription and metabolism in the liver of high-fat diet-fed C57BL/6 J mice. Mol Nutr Food Res 2011; 55: S173–S185.
Kwon EY, Shin SK, Cho YY, Jung UJ, Kim E, Park T et al. Time-course microarrays reveal early activation of the immune transcriptome and adipokine dysregulation leads to fibrosis in visceral adipose depots during diet-induced obesity. BMC Genomics 2012; 13: 450.
Fraulob JC, Ogg-Diamantino R, Fernandes-Santos C, Aguila MB, Mandarim-de-Lacerda CA . A mouse model of metabolic syndrome: insulin resistance, fatty liver and non-alcoholic fatty pancreas disease (NAFPD) in C57BL/6 mice fed a high fat diet. J Clin Biochem Nutr 2010; 46: 212–223.
Gregoire FM, Zhang Q, Smith SJ, Tong C, Ross D, Lopez H et al. Diet-induced obesity and hepatic gene expression alterations in C57BL/6 J and ICAM-1-deficient mice. Am J Physiol Endocrinol Metab 2002; 282: E703–E713.
Kim S, Sohn I, Ahn JI, Lee KH, Lee YS, Lee YS . Hepatic gene expression profiles in a long-term high-fat diet-induced obesity mouse model. Gene 2004; 340: 99–109.
Kleemann R, van Erk M, Verschuren L, van den Hoek AM, Koek M, Wielinga PY et al. Time-resolved and tissue-specific systems analysis of the pathogenesis of insulin resistance. PLoS One 2010; 5: e8817.
Radonjic M, de Haan JR, van Erk MJ, van Dijk KW, van den Berg SA, de Groot PJ et al. Genome-wide mRNA expression analysis of hepatic adaptation to high-fat diets reveals switch from an inflammatory to steatotic transcriptional program. PLoS One 2009; 4: e6646.
Huang, da W, Sherman BT, Lempicki RA . Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 2009; 4: 44–57.
Draghici S, Khatri P, Bhavsar P, Shah A, Krawetz SA, Tainsky MA . Onto-Tools, the toolkit of the modern biologist: Onto-Express, Onto-Compare, Onto-Design and Onto-Translate. Nucleic Acids Res 2003; 31: 3775–3781.
Al-Shahrour F, Diaz-Uriarte R, Dopazo J . FatiGO: a web tool for finding significant associations of Gene Ontology terms with groups of genes. Bioinformatics 2004; 20: 578–580.
Zeeberg BR, Feng W, Wang G, Wang MD, Fojo AT, Sunshine M et al. GoMiner: a resource for biological interpretation of genomic and proteomic data. Genome Biol 2003; 4: R28.
Kanehisa M, Goto S, Sato Y, Furumichi M, Tanabe M . KEGG for integration and interpretation of large-scale molecular data sets. Nucleic Acids Res 2012; 40: D109–D114.
Lee I, Blom UM, Wang PI, Shim JE, Marcotte EM . Prioritizing candidate disease genes by network-based boosting of genome-wide association data. Genome Res 2011; 21: 1109–1121.
Kim J, Choi M, Kim JR, Jin H, Kim VN, Cho KH . The co-regulation mechanism of transcription factors in the human gene regulatory network. Nucleic Acids Res 2012; 40: 8849–8861.
Gerstein MB, Kundaje A, Hariharan M, Landt SG, Yan KK, Cheng C et al. Architecture of the human regulatory network derived from ENCODE data. Nature 2012; 489: 91–100.
Fazekas D, Koltai M, Turei D, Modos D, Palfy M, Dul Z et al. SignaLink 2-a signaling pathway resource with multi-layered regulatory networks. BMC Syst Biol 2013; 7: 7.
Blake JA, Harris MA . The Gene Ontology (GO) project: structured vocabularies for molecular biology and their application to genome and expression analysis. Curr Protoc Bioinformatics 2008; Chapter 7, Unit 7.2.
Xu H, Barnes GT, Yang Q, Tan G, Yang D, Chou CJ et al. Chronic inflammation in fat plays a crucial role in the development of obesity-related insulin resistance. J Clin Invest 2003; 112: 1821–1830.
Patel PS, Buras ED, Balasubramanyam A . The role of the immune system in obesity and insulin resistance. J Obes 2013; 2013: 616193.
Sun B, Karin M . Obesity, inflammation, and liver cancer. J Hepatol 2012; 56: 704–713.
Kim JR, Shin D, Jung SH, Heslop-Harrison P, Cho KH . A design principle underlying the synchronization of oscillations in cellular systems. J Cell Sci 2010; 123: 537–543.
Kim TH, Kim J, Heslop-Harrison P, Cho KH . Evolutionary design principles and functional characteristics based on kingdom-specific network motifs. Bioinformatics 2011; 27: 245–251.
Consortium EP. An integrated encyclopedia of DNA elements in the human genome. Nature 2012; 489: 57–74.
Massrieh W, Derjuga A, Doualla-Bell F, Ku CY, Sanborn BM, Blank V . Regulation of the MAFF transcription factor by proinflammatory cytokines in myometrial cells. Biol Reprod 2006; 74: 699–705.
Moran JA, Dahl EL, Mulcahy RT . Differential induction of mafF, mafG and mafK expression by electrophile-response-element activators. Biochem J 2002; 361: 371–377.
Yamazaki H, Katsuoka F, Motohashi H, Engel JD, Yamamoto M . Embryonic lethality and fetal liver apoptosis in mice lacking all three small Maf proteins. Mol Cell Biol 2012; 32: 808–816.
Tuikkala J, Vahamaa H, Salmela P, Nevalainen OS, Aittokallio T . A multilevel layout algorithm for visualizing physical and genetic interaction networks, with emphasis on their modular organization. BioData Min 2012; 5: 2.
Smoot ME, Ono K, Ruscheinski J, Wang PL, Ideker T . Cytoscape 2.8: new features for data integration and network visualization. Bioinformatics 2011; 27: 431–432.
Newman ME . Modularity and community structure in networks. Proc Natl Acad Sci USA 2006; 103: 8577–8582.
Morris JH, Apeltsin L, Newman AM, Baumbach J, Wittkop T, Su G et al. clusterMaker: a multi-algorithm clustering plugin for Cytoscape. BMC Bioinformatics 2011; 12: 436.
Edgar R, Domrachev M, Lash AE . Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res 2002; 30: 207–210.
Kennedy AR, Pissios P, Otu H, Roberson R, Xue B, Asakura K et al. A high-fat, ketogenic diet induces a unique metabolic state in mice. Am J Physiol Endocrinol Metab 2007; 292: E1724–E1739.
Singh R, Mortazavi A, Telu KH, Nagarajan P, Lucas DM, Thomas-Ahner JM et al. Increasing the complexity of chromatin: functionally distinct roles for replication-dependent histone H2A isoforms in cell proliferation and carcinogenesis. Nucleic Acids Res 2013; 41: 9284–9295.
Wu RS, Bonner WM . Separation of basal histone synthesis from S-phase histone synthesis in dividing cells. Cell 1981; 27: 321–330.
Hu X, Zhang P, Xu Z, Chen H, Xie X . GPNMB enhances bone regeneration by promoting angiogenesis and osteogenesis: potential role for tissue engineering bone. J Cell Biochem 2013; 114: 2729–2737.
Tang SY, Herber RP, Ho SP, Alliston T . Matrix metalloproteinase-13 is required for osteocytic perilacunar remodeling and maintains bone fracture resistance. J Bone Miner Res 2012; 27: 1936–1950.
Arnott JA, Lambi AG, Mundy C, Hendesi H, Pixley RA, Owen TA et al. The role of connective tissue growth factor (CTGF/CCN2) in skeletogenesis. Crit Rev Eukaryot Gene Expr 2011; 21: 43–69.
Evans DS, Cailotto F, Parimi N, Valdes AM, Castano-Betancourt MC, Liu Y et al. Genome-wide association and functional studies identify a role for IGFBP3 in hip osteoarthritis. Ann Rheum Dis 2014; (pii) annrheumdis-2013-205020 e-pub ahead of print 13 June 2014 doi:10.1136/annrheumdis-2013-205020.
Zhao LJ, Jiang H, Papasian CJ, Maulik D, Drees B, Hamilton J et al. Correlation of obesity and osteoporosis: effect of fat mass on the determination of osteoporosis. J Bone Miner Res 2008; 23: 17–29.
Lv S, Wu L, Cheng P, Yu J, Zhang A, Zha J et al. Correlation of obesity and osteoporosis: effect of free fatty acids on bone marrow-derived mesenchymal stem cell differentiation. Exp Ther Med 2010; 1: 603–610.
McGregor RA, Kwon EY, Shin SK, Jung UJ, Kim E, Park JH et al. Time-course microarrays reveal modulation of developmental, lipid metabolism and immune gene networks in intrascapular brown adipose tissue during the development of diet-induced obesity. Int J Obes (Lond) 2013; 37: 1524–1531.
Barbatelli G, Murano I, Madsen L, Hao Q, Jimenez M, Kristiansen K et al. The emergence of cold-induced brown adipocytes in mouse white fat depots is determined predominantly by white to brown adipocyte transdifferentiation. J Physiol Endocrinol Metab 2010; 298: E1244–E1253.
Acknowledgements
We thank Jinmuk Kang, Bona Lee and Jinny Choe for their helpful discussions. This work was supported by the Bio-Synergy Research Project (NRF-2012M3A9C4048735) and NRF-2012R1A1A2007188 of the Ministry of Science, ICT and Future Planning through the National Research Foundation, Korea.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on International Journal of Obesity website
Supplementary information
Rights and permissions
About this article
Cite this article
Kim, J., Kwon, EY., Park, S. et al. Integrative systems analysis of diet-induced obesity identified a critical transition in the transcriptomes of the murine liver and epididymal white adipose tissue. Int J Obes 40, 338–345 (2016). https://doi.org/10.1038/ijo.2015.147
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ijo.2015.147
This article is cited by
-
Common dysregulated pathways in obese adipose tissue and atherosclerosis
Cardiovascular Diabetology (2016)