Abstract
Objective:
To investigate the impact of reduced adipocyte fatty acid-binding protein 4 (FABP4) in control of body weight, glucose and lipid homeostasis in diet-induced obese (DIO) mice.
Methods:
We applied RNA interference (RNAi) technology to generate FABP4 germline knockdown mice to investigate their metabolic phenotype.
Results:
RNAi-mediated knockdown reduced FABP4 mRNA expression and protein levels by almost 90% in adipocytes of standard chow-fed mice. In adipocytes of DIO mice, RNAi reduced FABP4 expression and protein levels by 70 and 80%, respectively. There was no increase in adipocyte FABP5 expression in FABP4 knockdown mice. The knockdown of FABP4 significantly increased body weight and fat mass in DIO mice. However, FABP4 knockdown did not affect plasma glucose and lipid homeostasis in DIO mice; nor did it improve their insulin sensitivity.
Conclusion:
Our data indicate that robust knockdown of FABP4 increases body weight and fat mass without improving glucose and lipid homeostasis in DIO mice.
Similar content being viewed by others
Introduction
Fatty acid-binding proteins (FABPs) are a family of conserved proteins that bind medium to long-chain fatty acids and are believed to coordinate lipid response in cells.1, 2, 3 Fatty acid-binding proteins not only facilitate long-chain fatty acid (LCFA) transport and metabolism, but also act as signaling molecules to sequester or distribute LCFA to regulate signaling pathways and gene expression. Recent studies demonstrated that dysregulated fatty acid metabolism and signaling have a crucial role in the etiology of metabolic syndrome. Both in vitro and in vivo studies showed that FABPs seem to be central in the regulation of fatty acid metabolism.1, 2, 3 However, the exact mechanisms by which FABPs regulate different biological functions are not well understood.
There are nine members of the FABP family, and each of them shows a distinct tissue expression pattern.1, 2, 3 Several lines of evidence indicate that adipocyte FABP (FABP4 or adipocyte protein 2 (aP2)), which is also expressed in macrophages, contributes to the development of several aspects of metabolic syndrome.4, 5, 6, 7 Mice with genetic disruption of FABP4 are protected against diet-induced insulin resistance despite the development of more severe obesity.4 Genetic disruption of FABP4 in obese diabetic mice (ob/ob) significantly reduces blood glucose levels and improves insulin sensitivity concomitant with increased body weight.7 These mice are also reported to have improved plasma lipid profile with the reduction in plasma triglyceride (TG) and cholesterol levels. Complete knockout of FABP4 or macrophage-specific FABP4 deficiency in apoE-deficient mice leads to marked protection against the development of atherosclerosis.5 Although FABP4 is the major FABP in adipocytes, another FABP protein (FABP5 or keratinocyte lipid binding protein (MAL1)) is also found in these cells.8 As genetic disruption of FABP4 significantly increases FABP5 expression, it was difficult to formally determine which aspects of the FABP4 knockout phenotype were due to the loss of FABP4 or upregulation of FABP5.4 Additional studies demonstrated that genetic disruption of FABP5 mildly improves insulin sensitivity in diet-induced obesity with a concomitant reduction in body weight.9 Furthermore, FABP5 knockout mice on an ob/ob background demonstrate a mild improvement in insulin sensitivity without an alteration in body weight. In contrast, transgenic mice with overexpression of FABP5 in adipocytes develop glucose intolerance.9 Genetic disruption of both FABP4 and FABP5 (double knockout) has a more profound metabolic effect in different animal models of insulin resistance than the ablation of only one isoform.10, 11 Double knockout mice show a marked improvement in insulin sensitivity and significant resistance to diet-induced obesity.11 Double knockout mice on an ob/ob background demonstrate improved insulin sensitivity and protection from hepatic steatosis.10 These studies indicate that FABP4 and FABP5 both have an important role in the development of insulin resistance. They also suggest that blocking the function of FABP4 and FABP5 could provide significant efficacy in reversing pathologies of nutrient metabolism. Indeed, a small molecule that has been shown to inhibit the binding of a fluorescently labeled fatty acid to FABP4 has been reported to reduce plasma glucose levels, improve insulin sensitivity and improve glucose tolerance without affecting body weight in ob/ob mice.12
Although total genetic disruption may generally serve as the best means to determine the function of a gene, it may not always be the best method to predict the biological outcome of pharmacologically inhibiting the protein product of that gene. For example, the statins, 3-hydroxy-3-methylglutaryl–coenzyme A reductase inhibitors, have been proven to be clinically safe and efficacious as antiatherosclerosis therapy, whereas genetic disruption of 3-hydroxy-3-methylglutaryl–coenzyme A reductase in mice results in embryonic lethality. Similarly, genetic disruption of FABP4 leads to a robust increase in FABP5 expression,4 which makes it difficult to determine whether the phenotype observed in FABP4 knockout mice is due to the loss of FABP4 or gain of FABP5 function.
Of late, RNA interference (RNAi) has become a powerful tool to understand gene function. Systemic delivery of RNAi in mice can effectively knockdown target gene in liver by different means.13, 14 However, knockdown of target gene in adipose tissue in vivo by RNAi remains challenging. To better predict the potential on-target effect of FABP4 pharmacologic inhibitors, we applied RNAi technology to generate a whole-body knockdown model of this FABP. In this study, we report the phenotypic characterization of this mouse line.
Materials and methods
FABP4 RNAi and shRNA
In preliminary studies, an RNAi sequence specific for mouse FABP4 was shown to effectively knockdown FABP4 expression in NIH3T3 cells. The sense sequence for the FABP4 RNAi contains 19 nucleotides: 5′-CACCGAGATTTCCTTCAAA-3′. To create a short hairpin RNA (shRNA), a nine-nucleotide loop (5′-TTCAAGAGA-3′) was placed to separate the sense and antisense sequences for RNAi.
Animals
The entire FABP4 shRNA described above was integrated into Rosa26 in 129S6 × C57BL/6 F1 mouse embryonic stem cells by TaconicArtemis (Cologne, Germany) as described before.15 Mice expressing FABP4 shRNA (shFABP4) were established and then backcrossed to C57BL/6NTac mice for six generations. At 5–6 weeks of age, these mice were housed in a group of 3–4 mice under standard housing conditions on a 12/12-h light and dark cycle. The mice were provided with ad libitum access to water and a standard chow diet (TEKLAD 7012: 13.4% kcal from fat; 3.41 kcal g−1; Harlan Teklad, Madison, WI, USA) until 8 weeks of age. At that time, half of the mice were switched to a high-fat diet (HFD; D12492: 60% kcal from fat, 5.24 kcal g−1; Research Diet, New Brunswick, NJ, USA) and continued for another 11 weeks. Body weight and food intake were monitored weekly. Body fat composition was analyzed using a Fat/Lean Mice Whole Body Magnetic Resonance Analyzer (Brucker, Milton, ON, Canada) at week 11 of the HFD. Oral glucose tolerance test was performed at week 11 on HFD (under 4-h fasted condition) with a bolus glucose dose of 2 g kg−1 of body weight. After a 2-week recovery period, an insulin tolerance test was performed with an intraperitoneal injection of 0.5 U kg−1 insulin (Humulin-R, Lilly, Indianapolis, IN, USA). Blood glucose levels were monitored every 20 min up to 120 min during the oral glucose tolerance test and insulin tolerance test. After another 2-week recovery, ad libitum fed or overnight fasted animals were killed and the liver, epididymal fat and plasma were collected for further analysis. All animal protocols used in these studies were approved by the Merck Research Laboratories Institutional Animal Care and Use Committee (Rahway, NJ, USA).
Plasma analysis
Glucose levels were analyzed from tail bleeds with a glucometer (Lifescan, Milpitas, CA, USA). Plasma insulin levels were measured with a commercial kit (ALPCO Diagnostics, Windham, NH, USA). Plasma TG and free fatty acid levels were also measured with commercial kits (TG (Cat: 11488872) and free fatty acid half micro test (Cat: 11383175), Roche, Branford, CT, USA).
RNA isolation and analysis
Liver and adipose tissue were homogenized with the Ultraspec RNA isolation kit (Biotecx, Houston, TX, USA). Total RNA was then prepared using the Qiagen RNAeasy 96 purification system (Qiagen, Valencia, CA, USA). The concentration of RNA was estimated from absorbance at 260 nm. The expression levels of specific mRNAs were quantified using quantitative fluorescent real-time PCR. The RNA was first reverse transcribed using the iSCript cDNA synthesis kit according to the protocol provided by the manufacturer (Bio-Rad, Hercules, CA, USA). The amplification of each target complementary DNA was then performed using TaqMan PCR Reagent Kits in the ABI Prism 7900 Sequence Detection System according to the protocols provided by the manufacturer (PE Applied Biosystems, Foster City, CA, USA). All primer/probe sets were selected from Applied Biosystems’ pre-developed TaqMan gene assays. The levels of mRNA were normalized to the amount of GAPDH mRNA detected in each sample.
Tissue lysis and FABP4 ELISA
Adipose tissue was homogenized at a concentration of 150 mg ml−1 in T-PER tissue protein extraction reagent (Pierce, Rockford, IL, USA) supplemented with Halt protease inhibitor cocktail (Pierce). Lysates were then centrifuged and cleared supernatants were collected. Protein concentration was determined using a Bradford assay (Pierce) and all samples were normalized to 50 μg ml−1. Samples were then analyzed for FABP4 using a mouse Adipocyte FABP ELISA kit (BioVendor, Candler, NC, USA) according to the manufacturer′s protocol.
Plasma desaturation index analysis
Fatty acid methyl esters were generated by boiling 0.1 ml of plasma in 3 ml of 5% methanolic HCl for 60 min. Methyl esters were extracted into hexane and then dried down under nitrogen. Samples were resuspended in methanol. All samples were analyzed using an Agilent 5975N-MSD equipped with an Agilent 6890N GC system (Agilent technologies, Santa Clara, CA, USA), and an HP-5MS capillary column (30 m × 0.25 mm × 0.25 μm). The temperature program was as follows: 120 °C initial, increase by 10 °C min−1 to 165 °C, hold for 30 min, increase by 5 °C min−1 to 210 °C, hold for 5 min, increase by 5 °C min−1 to 230 °C, hold for 7 min. The split ratio was 10:1 with a helium flow of 1 ml min−1. Elution times were as follows: palmitoleic acid methyl ester=15.8 min; palmitic acid methyl ester=16.2 min; oleic acid=18.3 min; and stearic acid methyl ester=18.8 min. The mass spectrometer was operated in the electron impact mode (70 eV). The instrument was operated in the scan mode from 50 to 320 m/z. Samples were analyzed in triplicate. A desaturation index was calculated as the ratio of monosaturated fatty acid vs saturated fatty acid levels.
Statistic analysis
Data are expressed as the mean±s.e.m. Statistical significance was determined by using the unpaired Student′s t-test. Statistical significance was assumed at P<0.05.
Results
Germline expression of FABP4 shRNA robustly knocks down expression of FABP4 in adipose tissue
To obtain insight into the potential outcome of pharmacologically inhibiting FABP4 function specifically, we generated a mouse germline knockdown model expressing FABP4 shRNA (shFABP4). Real-time PCR analysis showed that shFABP4 knocked down FABP4 gene expression in adipose tissue by 88% compared to wild-type (WT) mice on standard chow (12±2 vs 100±15 arbitrary units (AUs), respectively; Figure 1a). Wild-type diet-induced obese (DIO) mice that had been fed an HFD, showed no difference in adipose tissue FABP4 expression in comparison with standard chow-fed (SC) mice. In DIO mice, shFABP4 knocked down adipose tissue FABP4 expression by 70% compared with WT controls (31±3 vs 103±8 AU, respectively). The HFD did not alter FABP4 expression in WT mice compared with WT mice fed with SC. However, HFD shFABP4 mice had a 2.5-fold increase in FABP4 mRNA levels compared with shFABP4 mice fed with SC, thereby resulting in only a 70% net diminution in FABP4 mRNA expression in HFD rather than an 88% reduction in SC mice (Figure 1a). Constitutive genetic disruption of FABP4 has previously been reported to robustly induce FABP5 expression. In contrast, knockdown of FABP4 by shRNA did not affect FABP5 expression in SC or DIO mice (Figure 1b). The HFD feeding robustly increased FABP5 expression in adipose tissue by more than sixfold in WT mice and ninefold in shFABP4 mice (Figure 1b). To confirm that FABP4 protein levels were reduced in adipose tissue of shFABP4 mice, we performed an ELISA of adipose tissue lysates as described in the ‘Materials and methods’ section. Consistent with our gene expression analysis results, shFABP4 diminished adipose tissue FABP4 protein levels by 88% (0.12±0.02 vs 1±0.16 AU) in SC mice (Figure 1c), and by 77% (0.21±0.04 vs 0.9±0.16 AU) in DIO mice. There was no change in FABP4 protein levels between WT SC mice and WT DIO mice. However, DIO shFABP4 mice displayed a 1.8-fold elevation in adipose tissue FABP4 protein levels compared with SC shFABP4 mice. In sum, our data indicated that RNAi-mediated germline knockdown of FABP4 robustly reduced adipose tissue FABP4 gene expression and protein levels without inducing FABP5 expression.
Germline expression of fatty acid-binding protein 4 (FABP4) short hairpin RNA (shRNA) (shFABP4) robustly knocks down FABP4 in adipose tissue without affecting FABP5 expression. Gene expression analysis was performed in adipose tissue from mice on standard chow (SC) and high-fat diet (HFD). Real-time PCR analysis of (a) FABP4 and (b) FABP5 expression relative to wild-type SC mice (16–20 animals per group). (c) ELISA analysis of FABP4 protein levels in adipose tissue relative to wild-type SC mice (16–20 animals per group). Expression levels in wild-type SC mice were artificially set to 100. Open bar and filled bar represents gene expression levels in wild-type mice and in shFABP4 mice, respectively. *Represents a significant difference between wild-type and shFABP4 mice in either SC or HFD mice (P<0.001).
Knockdown of FABP4 significantly increases body weight and whole-body fat mass in response to HFD feeding
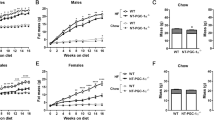
To characterize the functional impact of FABP4 knockdown, we analyzed the body weight of SC and HFD mice. At week 10, male shFABP4 mice fed SC displayed a significant reduction in body weight gain compared with WT mice on the same diet (21.4±1.6 vs 19.8±2 g, respectively; Figure 2a); this difference was maintained throughout the remainder of the 20 week study. Half of the mice were switched to HFD at 9 weeks of age. After 7 weeks of high fat feeding, male shFABP4 mice showed a significant increase in body weight in comparison with WT mice on the same HFD (42.4±0.9 vs 39±1 g, respectively); this difference continued to grow throughout the length of study (51.2±0.6 vs 45.8±0.7 g at 20 weeks, respectively). Female shFABP4 mice showed a similar relative increase in body weight on HFD and reduction in body weight gain on SC vs WT mice (data not shown). In comparison to WT mice, shFABP4 mice provided the HFD displayed a significant increase in weekly food intake starting from 13 week (data not shown), which is 4 weeks after the commencement of HFD feeding (Figure 2b). There was no significant difference in food intake between shFABP4 and WT mice on SC.
RNA interference (RNAi)-mediated knock down of fatty acid-binding protein 4 (FABP4) significantly increases body weight and fat mass in mice on high-fat diet (HFD). (a) Body weight and (b) weekly food intake per mouse on HFD or standard chow (SC) diet (16–20 animals per group; P<0.01). Open square and filled square represent wild-type mice and shFABP4 mice on HFD; open triangle and filled triangle represent wild-type mice and shFABP4 mice on SC diet, respectively. (c) Nuclear magnetic resonance (NMR) analysis of body fat composition presented as percentage to body weight (16–20 animals per group; P<0.001). Open bar and filled bar represents percentage of fat mass in wild-type mice and in shFABP4 mice, respectively. *Represents a significant difference between wild-type and shFABP4 mice in either SC or HFD mice.
We examined the whole body fat mass of our different mouse cohorts using the quantitative nuclear magnetic resonance methodology described in the ‘Materials and methods’ section. There was no difference in the body fat content between shFABP4 and WT mice on SC. However, as expected, WT mice fed the HFD demonstrated an almost fourfold increase in fat mass compared with SC mice (46.3±0.8 vs 12.4±0.8%, respectively; Figure 2c). Furthermore, shFABP4 mice on HFD showed a marked increase in fat mass vs WT HFD mice (52.1±1.1 vs 46.3±0.8%, respectively; Figure 2c). A different set of mice (n=7–10 per group) was studied at Mouse Clinical Institute (MCI) (Parc d'Innovation, France) with consistent results in body weight and fat mass. Taken together, our data indicated that knockdown of FABP4 robustly increases body weight and fat mass in HFD mice.
Knocking down FABP4 does not affect glucose tolerance or insulin sensitivity
To investigate the potential physiological effects of FABP4 knockdown, we analyzed several key plasma metabolic parameters in ad libitum fed or overnight fasted mice. As shown in Table 1, both WT and shFABP4 HFD mice showed a 27-fold increase in ambient plasma insulin levels compared with SC mice. In contrast, plasma glucose, TG and free fatty acid levels did not differ in HFD vs SC mice. The knockdown of FABP4 did not significantly affect plasma insulin, glucose, TG or free fatty acid levels in ad libitum fed SC or HFD mice. Nor did it have a notable impact on fasting plasma insulin, glucose, TG or free fatty acid levels in SC or HFD mice (Table 2). As knocking out FABP4 has been shown to improve glucose tolerance and insulin sensitivity in HFD mice, we performed oral glucose tolerance test and insulin tolerance test. As shown in Figure 3a, knockdown of FABP4 had no impact on glucose tolerance in HFD and SC mice. In addition, knockdown of FABP4 by shRNA did not significantly alter insulin sensitivity in HFD and SC mice as measured by their response to an insulin challenge (Figure 3b). These results were confirmed by an independent study using a different cohort of mice. These data demonstrated that robust knockdown of FABP4 had no effect on glucose homeostasis or insulin action in DIO mice.
Effect of fatty acid-binding protein 4 (FABP4) knockdown on glucose tolerance and insulin sensitivity in diet-induced obese mice. (a) Oral glucose tolerance test and (b) insulin tolerance test were performed in high-fat diet (HFD) and standard chow (SC) mice (8–10 animals per group, P>0.05). A bolus of glucose (2 g kg−1 of body weight) was dosed orally. Insulin (0.5 U kg−1 of body weight) was administered by intraperitoneal (IP) injection. Glucose levels were monitored at 20, 40, 60, 90 and 120 min by tail bleed method. Filled triangle, filled square, open triangle and open square represent HFD wild-type (WT), HFD shFABP4, SC WT and SC shFABP4 mice, respectively.
Effects on plasma LCFA desaturation index by knockdown of FABP4 in HFD mice
Mice with genetic disruption of both FABP4 and FABP5 are protected against the development of fatty liver, which have significantly reduced liver TGs.11 Further analysis demonstrated that hepatic stearoyl coenzyme A desaturase 1 (SCD1) levels and activity were significantly reduced. Stearoyl CoA desaturase 1 has a key role in regulating hepatic lipogenesis, and inhibition of this enzyme lowers liver TG content.16 In addition, the level of liver SCD1 expression has been shown to regulate the plasma LCFA desaturation index.16 Although the mechanism by which FABP4 and FABP5 regulate hepatic SCD1 expression is not currently understood, this association could provide a potential biomarker for a drug development program targeting FABP4 and FABP5.
We observed no changes in hepatic TG levels in shFABP4 mice compared with WT mice on the HFD (data not shown). To investigate the effect of shFABP4 on plasma LCFA desaturation index, we used gas chromatography–mass spectrometry to analyze the saturation levels of plasma LCFA in our different mouse groups and subsequently calculate a desaturation index as described in the ‘Materials and methods’ section. In ad libitum fed WT mice, HFD tended to reduce the plasma C18:1/C18:0 desaturation index (−30%) and significantly reduced the plasma C16:1/C16:0 desaturation index (−40%) compared with SC mice (Figures 4a, b). In comparison with shFABP4 mice on SC, shFABP4 mice on HFD showed significantly reduced plasma C18:1/C18:0 and C16:1/C16:0 indices (59 and 65%, respectively). In comparison with WT mice, shFABP4 mice on SC did not show a change in their plasma LCFA desaturation indices, whereas shFABP4 mice on HFD tended to demonstrate a modest reduction in C18:1/C18:0 and C16:1/C16:0 desaturation indices (34 and 27%, respectively). Subsequently, we analyzed hepatic SCD1 gene expression in our various mouse cohorts. In comparison with WT mice fed an HFD, shFABP4 mice tended to show a reduction (−29%) in liver SCD1 expression, whereas shFABP4 mice on SC showed no change in SCD1 expression (Figure 4c). These data indicated that shRNA-mediated knockdown of FABP4 only minimally reduced the plasma LCFA desaturation indices and hepatic SCD1 expression in HFD mice.
Effect of fatty acid-binding protein 4 (FABP4) knockdown on the plasma long chain fatty acid (LCFA) desaturation index. In ad libitum fed mice, plasma C18 and C16 fatty acid desaturation index was measured for the ratio of monounsaturated FFA vs saturated FFA by gas chromatography–mass spectrometry (GC/MS) as described in Materials and Methods section. (a) C18:1/C18:0 and (b) C16:1/C16:0 (8–10 animals per group, P>0.05). (c) Real-time PCR analysis of liver stearoyl coenzyme A desaturase 1 (SCD1) mRNA expression (8–10 animals per group, P>0.05). Open bar and filled bar represent wild-type mice and shFABP4 mice, respectively.
Effects of FABP4 knockdown on adipose tissue inflammation in HFD mice
Recent studies have shown that FABP4 is expressed in adipocytes and macrophages and its action in either cell types is critical for the impact of FABP4 on systematic metabolism.17 To investigate the knockdown of FABP4 in macrophages, we analyzed FABP4 expression in the liver because it contains macrophages but not adipocytes and FABP4 is not expressed in hepatocytes. TaqMan analysis showed that there was an 80 or 83% reduction of FABP4 expression in SC or HFD shFABP4 liver compared with WT mice, respectively (Figure 5a). We could not detect the fat-specific gene, leptin, in those livers, suggesting few adipocytes in the liver samples (Figure 5a). Liver samples expressed several macrophage-specific marker genes, including CD68, F4/80, MIP-1 and ADAM8, thus indicating the presence of macrophages (Figure 5a).18 Indeed, HFD mice demonstrated significant increases in CD68 and MIP-1 expressions and trends toward increased F4/80 and ADAM8 compared with SC mice, suggesting increased hepatic macrophage infiltration in HFD mice. Taken together, these data support the conclusion that knockdown of FABP4 in liver is mainly due to knockdown of FABP4 in macrophages.
Effect of fatty acid-binding protein 4 (FABP4) knockdown on macrophages and adipose tissue inflammation. Gene expression analysis was performed in liver and adipose tissue from mice on standard chow (SC) and high-fat diet (HFD). (a) Real-time PCR analysis of liver samples for FABP4, leptin and macrophage marker genes: CD68 (macrosialin), F4/80 (macrophage-restricted surface glycoprotein), MIP-1 (macrophage inflammatory protein-1) and ADAM8 (disintegrin-like metalloproteinase). (b) Real-time PCR analysis of adipose tissues for leptin and macrophage marker genes: CD68, F4/80, MIP-1, MAC-1 (macrophage antigen-1) and ADAM8. Data represented as fold changes relative to SC wild-type WT controls. Open bar and filled bar represents gene expression levels in WT mice and in shFABP4 mice, respectively. *Represents a significant difference between WT and shFABP4 mice in either SC or HFD mice (P<0.05). ND, not detectable.
Several lines of evidence indicate that obesity is associated with a state of chronic, low-grade inflammation that contributes to insulin resistance and type 2 diabetes.19 To investigate the impact of FABP4 knockdown on adipose tissue inflammation, we analyzed the expression of macrophage markers in the adipose tissue. Leptin is highly expressed in adipose tissue and there was a significant increase in leptin expression in HFD mice compared with SC mice (Figure 5b). The macrophage markers CD68, F4/80, MIP-1, MAC-1 and ADAM8 are expressed in adipose tissue of both SC and HFD mice. Most of these macrophage marker genes were robustly upregulated in HFD adipose tissue, suggesting significant macrophage infiltration in adipose tissue of HFD mice (Figure 5b). In HFD mice, knockdown of FABP4 significantly increased the expression of all the macrophage markers we tested, indicating increased adipose tissue inflammation in shFABP4 HFD mice. We did not observe increases in the expression of these macrophage markers in SC shFABP4 adipose tissue compared with SC WT mice (Figure 5b). These data suggest that knockdown of FABP4 increases adipose inflammation but does not induce more severe insulin resistance in HFD mice.
Discussion
Fatty acid-binding proteins are a family of proteins that bind medium- and long-chain fatty acids and have been implicated in intracellular fatty acid trafficking.1, 3 The two major FABPs expressed in adipose tissue are FABP4 and FABP5; they are also expressed in macrophage. In adipocytes, FABP4 is more abundant than FABP5, making up as much as 1% of their total soluble protein.8 The exact biological functions of FABP4 and FABP5 are not entirely understood; however, genetic disruption of FABP4 and FABP5 has been shown to result in improved metabolic phenotypes in obese insulin-resistant and diabetic mouse models.4, 11, 9, 7 These studies demonstrate a role for FABP4 and FABP5 in regulating insulin sensitivity and glucose homeostasis. Such data have triggered interest in developing pharmacological inhibitors of FABP4/5 to treat metabolic disorders.
Although knockout mouse models can provide a wealth of information regarding the physiological impact of complete absence of a molecular target, they are often unable to clearly indicate whether partial inhibition of a given target's intrinsic activity will also produce similar effects. Such information becomes more critical in the context of trying to identify a new drug target, as clinically useful small molecules are unlikely to completely inhibit the function of their target. To investigate whether a partial loss of FABP4 function will improve the pathophysiology observed in metabolic syndrome, we created a mouse line expressing an shRNA to FABP4. These mice displayed greatly reduced adipose FABP4 mRNA and protein levels relative to WT mice. In comparison with WT mice, our shFABP4 mice gained significantly more weight on HFD, which is consistent with the previously described phenotype of FABP4 knockout mice.3 As FABP4 knockout mice demonstrated an increase in FABP5 levels, this augmentation was initially thought to be responsible for the increase in adiposity seen in these animals. However, forced overexpression of FABP5 was subsequently observed to be body weight neutral in mice.4 In the shFABP4 mice described in this study, expression of FABP5 in adipose tissue was unchanged, yet these animals weighed more than WT mice. These data lead us to conclude that FABP5 did not contribute to the increase in body weight observed in shFABP4 mice, a deduction in line with the above FABP5 knockout mouse results. These data also hint at a possible untoward effect of FABP4-specific inhibitors on body weight when provided in a milieu of overnutrition.
Our results with shFABP4 mice clearly indicate that body weight is altered in a diet-dependent manner. Although these mice gained more weight than WT mice on an HFD, they showed a significant reduction in body weight compared with WT mice on SC diet containing very low levels of dietary fat. Comparing primary cultured adipocytes from WT and FABP4 knockout mice, the Bernlohr20 laboratory demonstrated that disruption of FABP4 did not affect the rate of fatty acid uptake or esterification. Furthermore, this group showed that disruption of FABP4 expression reduces basal and isoproterenol-stimulated lipolysis in primary adipocytes.20 They concluded that FABP4 functions as a lipid chaperone to facilitate the movement of fatty acids out of the cells. This function of FABP4 could provide a potential mechanism to explain the unique body weight phenotypes of shFABP4 mice fed different diets. When there is an excessive lipid load, as in HFD feeding, knockdown of FABP4 may reduce the removal of fatty acids out of adipocytes, thereby leading to greater storage of fat in adipose tissue and increased whole body fat mass and weight. As HFD shFABP4 mice could store more energy in the fat tissue, these animals could eat more to maintain daily energy utilization. Indeed, HFD shFABP4 mice had higher daily food intake than HFD WT mice. In contrast, during a low lipid load, such as that provided by an SC diet, adipocytes will only need to store very limited amounts of lipids and should therefore require only a relatively small amount of FABP4 protein to be fully functional. This proposition is supported by the FABP4 gene expression in this study. First, HFD did not induce FABP4 expression in WT mice compared with SC-fed mice, suggesting that excess FABP4 protein is available to handle the low lipid load in SC mice. Second, FABP4 proteins were induced in HFD shFABP4 mice compared with SC shFABP4 mice, thereby suggesting that the level of FABP4 protein in shFABP4 SC mice may be enough to handle their low lipid load. Therefore, when lipid load is increased in HFD shFABP4 mice, a compensatory machinery is activated to provide a necessary boost in FABP4 levels. Nevertheless, it is not clear why diminishing adipose FABP4 levels reduce body weight gain in mice fed an SC diet.
Complete ablation of FABP4, despite causing an increase in adiposity, has been shown to result in resistance to HFD-induced metabolic dysfunction.3 In contrast, in our shFABP4 HFD mice, we were able to reduce adipose FABP4 protein levels by 77%, but still did not observe significant improvements in insulin sensitivity or glucose tolerance. Further analysis indicated that adipose tissue inflammation was increased in shFABP4 HFD mice compared with HFD WT mice. Although shFABP4 HFD mice were more obese and had more adipose tissue inflammation, these mice did not develop more severe insulin resistance compared with WT HFD mice. These data suggest that the beneficial impact in systematic metabolism by knockdown of FABP4, if any, may be concealed by increased adiposity and adipose tissue inflammation. Importantly, the reduction in FABP4 levels by shFABP4 was enough to increase the body weight on HFD without inducing FABP5 expression. Such data do not seem to support the use of FABP4-specific inhibitors as means to reverse derangements in nutrient metabolism brought about by overnutrition. However, Furuhashi et al.1 have recently provided the first report of the beneficial in vivo effects of a putative FABP4 small molecule inhibitor, BMS309403. They observed that chronic treatment with BMS309403 reduces elevated glucose and insulin levels, and mitigates glucose intolerance and insulin resistance in obese diabetic ob/ob mice.12 In addition, this small molecule did not seem to significantly affect body weight. Although such data might suggest that pharmacological inhibition of FABP4 function may result in desirable antidiabetic outcomes, the authors did not provide any data demonstrating the isoform specificity of BMS309403 in these animals, thereby leaving open the possibility that the beneficial metabolic effects they observed were due to dual inhibition of FABP4 and FABP5. Therefore, on the basis of previously described results with FABP4/5 double knockout mice,11 the above pharmacological study12 and the FABP4 knockdown mouse data we provided in this study, we suggest that a dual FABP4/5 inhibitor might prove most efficacious as an antidiabetic therapeutic agent with weight neutral or reducing effects. No doubt, an extensive medicinal chemistry effort and a broad array of in vitro and in vivo studies will need to be undertaken to validate this proposition and bring forth an effective new therapy for the treatment of type 2 diabetes patients.
References
Furuhashi M, Hotamisligil GS . Fatty acid-binding proteins: role in metabolic diseases and potential as drug targets. Nat Rev Drug Discov 2008; 7: 489–503.
Haunerland NH, Spener F . Fatty acid-binding proteins—insights from genetic manipulations. Prog Lipid Res 2004; 43: 328–349.
Hertzel AV, Bernlohr DA . The mammalian fatty acid-binding protein multigene family: molecular and genetic insights into function. Trends Endocrinol Metab 2000; 11: 175–180.
Hotamisligil GS, Johnson RS, Distel RJ, Ellis R, Papaioannou VE, Spiegelman BM . Uncoupling of obesity from insulin resistance through a targeted mutation in aP2, the adipocyte fatty acid binding protein. Science 1996; 274: 1377–1379.
Makowski L, Boord JB, Maeda K, Babaev VR, Uysal KT, Morgan MA et al. Lack of macrophage fatty-acid-binding protein aP2 protects mice deficient in apolipoprotein E against atherosclerosis. Nat Med 2001; 7: 699–705.
Tuncman G, Erbay E, Hom X, De Vivo I, Campos H, Rimm EB et al. A genetic variant at the fatty acid-binding protein aP2 locus reduces the risk for hypertriglyceridemia, type 2 diabetes, and cardiovascular disease. Proc Natl Acad Sci USA 2006; 103: 6970–6975.
Uysal KT, Scheja L, Wiesbrock SM, Bonner-Weir S, Hotamisligil GS . Improved glucose and lipid metabolism in genetically obese mice lacking aP2. Endocrinology 2000; 141: 3388–3396.
Simpson MA, LiCata VJ, Ribarik Coe N, Bernlohr DA . Biochemical and biophysical analysis of the intracellular lipid binding proteins of adipocytes. Mol Cell Biochem 1999; 192: 33–40.
Maeda K, Uysal KT, Makowski L, Gorgun CZ, Atsumi G, Parker RA et al. Role of the fatty acid binding protein mal1 in obesity and insulin resistance. Diabetes 2003; 52: 300–307.
Cao H, Maeda K, Gorgun CZ, Kim HJ, Park SY, Shulman GI et al. Regulation of metabolic responses by adipocyte/macrophage fatty acid-binding proteins in leptin-deficient mice. Diabetes 2006; 55: 1915–1922.
Maeda K, Cao H, Kono K, Gorgun CZ, Furuhashi M, Uysal KT et al. Adipocyte/macrophage fatty acid binding proteins control integrated metabolic responses in obesity and diabetes. Cell Metab 2005; 1: 107–119.
Furuhashi M, Tuncman G, Gorgun CZ, Makowski L, Atsumi G, Vaillancourt E et al. Treatment of diabetes and atherosclerosis by inhibiting fatty-acid-binding protein aP2. Nature 2007; 447: 959–965.
Aigner A . Cellular delivery in vivo of siRNA-based therapeutics. Curr Pharm Des 2008; 14: 3603–3619.
Yang R, Wilcox DM, Haasch DL, Jung PM, Nguyen PT, Voorbach MJ et al. Liver-specific knockdown of JNK1 up-regulates proliferator-activated receptor gamma coactivator 1 beta and increases plasma triglyceride despite reduced glucose and insulin levels in diet-induced obese mice. J Biol Chem 2007; 282: 22765–22774.
Seibler J, Kuter-Luks B, Kern H, Streu S, Plum L, Mauer J et al. Single copy shRNA configuration for ubiquitous gene knockdown in mice. Nucleic Acids Res 2005; 33: e67.
Jiang G, Li Z, Liu F, Ellsworth K, Dallas-Yang Q, Wu M et al. Prevention of obesity in mice by antisense oligonucleotide inhibitors of stearoyl-CoA desaturase-1. J Clin Invest 2005; 115: 1030–1038.
Furuhashi M, Fucho R, Gorgun CZ, Tuncman G, Cao H, Hotamisligil GS . Adipocyte/macrophage fatty acid-binding proteins contribute to metabolic deterioration through actions in both macrophages and adipocytes in mice. J Clin Invest 2008; 118: 2640–2650.
Xu H, Barnes GT, Yang Q, Tan G, Yang D, Chou CJ et al. Chronic inflammation in fat plays a crucial role in the development of obesity-related insulin resistance. J Clin Invest 2003; 112: 1821–1830.
Hotamisligil GS . Inflammation and metabolic disorders. Nature 2006; 444: 860–867.
Coe NR, Simpson MA, Bernlohr DA . Targeted disruption of the adipocyte lipid-binding protein (aP2 protein) gene impairs fat cell lipolysis and increases cellular fatty acid levels. J Lipid Res 1999; 40: 967–972.
Acknowledgements
We thank Drs C Li, T Akiyama, and C Lerner for helpful suggestions and support in this study.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies the paper on International Journal of Obesity website
Supplementary information
Rights and permissions
This work is licensed under the Creative Commons Attribution-NonCommercial-No Derivative Works 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/3.0/
About this article
Cite this article
Yang, R., Castriota, G., Chen, Y. et al. RNAi-mediated germline knockdown of FABP4 increases body weight but does not improve the deranged nutrient metabolism of diet-induced obese mice. Int J Obes 35, 217–225 (2011). https://doi.org/10.1038/ijo.2010.128
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ijo.2010.128
Keywords
This article is cited by
-
A Novel Missense SNP in the Fatty Acid-Binding Protein 4 (FABP4) Gene is Associated with Growth Traits in Karakul and Awassi Sheep
Biochemical Genetics (2023)
-
Fetal macrosomia in a Hispanic/Latinx predominant cohort and altered expressions of genes related to placental lipid transport and metabolism
International Journal of Obesity (2020)
-
Different expression of lipid metabolism-related genes in Shandong black cattle and Luxi cattle based on transcriptome analysis
Scientific Reports (2020)
-
RNA-seq analysis of the kidneys of broiler chickens fed diets containing different concentrations of calcium
Scientific Reports (2017)
-
A-FABP mediates adaptive thermogenesis by promoting intracellular activation of thyroid hormones in brown adipocytes
Nature Communications (2017)