Abstract
Prospective studies have reported that circulating cholesterol abnormalities are predictive of hypertension. As a test of the causal influence of circulating cholesterols on hypertension, we used a meta-based Mendelian randomization analysis to determine whether the apolipoprotein E (ApoE) gene polymorphism ɛ2/ɛ3/ɛ4 related to cholesterol changes is associated with hypertension. Data were available from 17 study populations, encompassing 2896 hypertensive patients and 2898 controls. A random effects model was applied irrespective of a between-study heterogeneity, and publication bias was examined using a funnel plot and Egger’s test. An overall comparison of the ApoE gene alleles ɛ4 with ɛ3 yielded a significant 81% increased risk for hypertension (95% confidence interval (95% CI): 1.41–2.32; P<0.0005). Restricting the analysis to populations of Asian descent resulted in a 1.87-times higher likelihood of developing hypertension (95% CI: 1.41–2.32; P<0.0005). Compared with ɛ3 carriers, ɛ4 allele carriers had significantly higher levels of total cholesterol (standardized mean difference (SMD)=0.39; 95% CI: 0.2–0.57; P<0.0005) and low-density lipoprotein cholesterol (SMD=0.43; 95% CI: 0.23–0.64; P<0.0005). The predicted odds ratio (OR) for a 1 mmol l−1 increase in total cholesterol was 4.58 (95% CI: 1.83–67.21) when all qualified studies were included and 4.98 (95% CI: 1.94–76.36) for Asian-descent populations. Similarly, the predicted OR for a 1-mmol l−1 increase of low-density lipoprotein cholesterol was 3.97 (95% CI: 1.71–43.48) in all populations and 4.29 (95% CI: 1.81–43.38) in Asian-descent populations. Taken together, our findings suggest a casual influence of high circulating total cholesterol and low-density lipoprotein cholesterol levels on the development of hypertension.
Similar content being viewed by others
Introduction
Prospective studies have generally reported that circulating lipid abnormalities are predictive of the subsequent development of hypertension.1, 2, 3 For example, a top-to-bottom fifth comparison of total cholesterol distribution yielded a relative multivariable-adjusted hypertension risk of 1.24 (95% confidence interval (95% CI): 1.01–1.53) in men,3 and age- and multivariable-adjusted risks of 1.21 (95% CI: 1.11–1.33) and 1.12 (95% CI: 1.02–1.23) in women.2 However, doubts about this claim have been raised by the observation that in women, the risk is considerably lower after multivariable adjustment, suggesting that the value is susceptible to confounding. Moreover, in prospective studies, the association between an exposure and disease is subject to Berkson's bias if the subject selection is based on an exposure-disease risk combination.4, 5 Furthermore, reverse causation might obscure this relationship, because obesity, an established risk factor for hypertension, may exert a less favorable influence on lipid levels. Consequently, understanding the influence of circulating lipids on hypertension requires additional studies to provide robust insights into the causal association between the intermediate phenotype and disease.
To minimize residual confounding factors and reverse causation, and to make causal inferences from observational epidemiology, an advisable approach is to apply Mendelian randomization with genetic variation as a proxy for a directly measured risk factor, because genotype distributions are believed to be independent of confounders and to be unmodified by disease processes.6 The rationale of the Mendelian randomization is to establish causality between the factor influenced by a polymorphism and a disease outcome via the joint polymorphism-disease and polymorphism-phenotype associations.7 Therefore, the different sustained levels of circulating lipids, such as total cholesterol and high- and low-density lipoprotein cholesterol, should translate into a differential risk of hypertension, if a causal link exists.
Polymorphisms of the apolipoprotein E (ApoE) gene have been studied extensively in the context of a variety of metabolic dyslipidemias and clinical endpoints, such as hypercholesterolemia,8, 9 Alzheimer's disease,10 stroke11 and hypertension.12 In particular, the ApoE gene polymorphism ɛ2/ɛ3/ɛ4 is of significant interest, because two recent meta-analyses consistently suggested that ɛ4 allele carriers were at increased risk for hypertension, and that this effect was more pronounced in Chinese populations.13, 14 Additionally, this polymorphism was reportedly linked to changes in cholesterol levels15, 16, 17, 18 and therefore should be associated with hypertension if a causality exists between cholesterol levels and hypertension.
Given the insufficient sample sizes of the individual studies that associated the ApoE gene ɛ2/ɛ3/ɛ4 polymorphism with circulating cholesterol, hypertension, or both, we decided to evaluate this association using a meta-analysis that aimed to test the causal influence of the circulating total cholesterol and high- and low-density lipoprotein cholesterol levels on hypertension.
Methods
Search strategy for the identification of studies
We identified the published articles through the PubMed and EMBASE search engines, as well as through the China Biological Medicine (http://sinomed.imicams.ac.cn/index.jsp) and Wanfang (http://www.wanfangdata.com.cn) databases before 18 January 2011, using the Boolean combination of subject terms (apolipoprotein E or ApoE or Apo E) and (hypertension or blood pressure) and (polymorphism or allele or genotype or variant or variation). The articles selected were restricted to human populations (other than families) and articles written in either English or Chinese. The full text of the retrieved articles was scrutinized to determine whether information on the topic of interest was included. The reference lists of the retrieved articles and systematic reviews were also checked for articles that had not been identified in the initial search. For studies involving more than one geographic or ethnic group, each group was treated separately.
Inclusion/exclusion criteria
Articles were considered qualified for this meta-analysis if they examined the hypothesis that the ApoE gene polymorphism ɛ2/ɛ3/ɛ4 is associated with hypertension, circulating cholesterols, or both; if they followed a case–control, nested case–control, or cross-sectional study design; and if they provided sufficient information on the ɛ2/ɛ3/ɛ4 genotype counts in hypertensive patients and controls for determining an estimate of odds ratio (OR) and the corresponding 95% CI, or sufficient information on circulating total cholesterol and high- and low-density lipoprotein cholesterol levels across the ɛ2/ɛ3/ɛ4 alleles for determining the standardized mean difference (SMD) and its 95% CI.
Hypertension was defined as systolic blood pressure equal to or greater than 140 mm Hg, diastolic blood pressure equal to or greater than 90 mm Hg, or previous treatment with antihypertensive drugs. Circulating cholesterols were measured using standard methods. Studies evaluating secondary hypertension or other types of monogenic hypertension were excluded. In cases in which there were multiple articles from the same study population (in whole or in part), the most complete and recent results were used.
Extracted information
The following data were extracted from each qualified study: the first author’s last name, the publication date, the population’s ethnicity, the baseline characteristics of the study population (including age, gender and body mass index), the ApoE ɛ2/ɛ3/ɛ4 genotype counts in patients and controls, and the circulating total cholesterol and high- and low-density lipoprotein cholesterol levels across the ɛ2 (ɛ2/2+ɛ2/3), ɛ3 (ɛ3/3) and ɛ4 (ɛ3/4+ɛ4/4) allele carriers. All cholesterol units were standardized to millimole per liter. For consistency, quantitative variables expressed as the mean±s.e. were converted to the mean±s.d. Data extraction and study quality were assessed in duplicate.
Statistical analysis
We used the random effects model and the DerSimonian and Laird method to combine the individual effect size estimates, and the estimate of heterogeneity was determined using the Mantel–Haenszel model.19 The unadjusted OR and SMD, and the corresponding 95% CI were used to compare genetic contrasts between patients and controls, as well as to compare the circulating cholesterol levels between ɛ3 allele carriers and carriers of the other ApoE alleles.
The between-study heterogeneity was assessed using the inconsistency index I2 statistic (ranging from 0 to 100%), which is defined as the percentage of the observed between-study variability, which is due to heterogeneity rather than chance. In this method, higher values suggest the existence of heterogeneity.20, 21 The funnel plots and Egger regression asymmetry test were used to assess publication bias. Egger’s test can detect funnel plot asymmetry by determining whether the intercept deviates significantly from zero in a regression of the standardized effect estimates against their precision.
It is widely accepted that genetic association studies are more closely relevant to randomized trials than other types of epidemiological studies owing to Mendel's second law of independent assortment of alleles, which states that assortment of alleles should be independent of environmental or behavioral factors.22, 23 In the Mendelian randomization analysis, the risk estimate was calculated from the ratio of the coefficient of the association between the polymorphism and disease to that of the association between the polymorphism and the assessed circulating cholesterol levels as a reflection of the potential causal effect of high circulating cholesterol levels on the hypertension risk.
A probability less than 0.05 was judged to be significant except for the I2 and Egger’s statistics, for which a significance level of less than 0.1 was chosen. The data management and statistical analyses were performed using STATA software, version 11.0 (StataCorp LP, College Station, TX, USA) for Windows.
Results
Search results and study characteristics
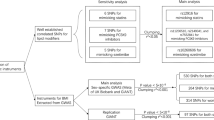
After our two recently published meta-analyses, which associated the ApoE gene polymorphism ɛ2/ɛ3/ɛ4 with hypertension using the results from English-language literature13 and from Chinese populations,14 we focused on whether circulating cholesterol levels related to this polymorphism are causal factors in hypertension, drawing from published studies in English and Chinese. In this study, we only assessed ɛ2/ɛ3/ɛ4 allelic association, so one study that provided combined genotype counts was removed.12 The detailed selection process is presented in Figure 1. A total of 17 articles,17, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39 involving 19 study populations with 2896 hypertensive patients and 2898 healthy controls, were analyzed. The baseline characteristics of all the qualified studies was mirrored in the above two meta-analyses.13, 14
A total of 7 of the 19 studies provided information regarding the association between the polymorphism and cholesterol. The distribution of total cholesterol and high- and low-density lipoprotein cholesterol levels across the ɛ2 (ɛ2/2+ɛ2/3), ɛ3 (ɛ3/3) and ɛ4 (ɛ3/4+ɛ4/4) alleles is summarized in Table 1. We must emphasize that, as suggested by Kesäniemi et al.,40 because of the opposite net effect between the ApoE ɛ2 and ɛ4 alleles, genotype ɛ2/4 should be excluded from the genotype–phenotype analysis.
Association of the ApoE gene alleles with hypertension
As shown in Figure 2a, a comparison of the ɛ2 vs. the ɛ3 ApoE allele indicated only a nonsignificantly (2%) increased risk for hypertension (OR=1.02; 95% CI: 0.83–1.26; P=0.834). This observation was accompanied by moderate evidence of between-study heterogeneity (I2=44.2%; P=0.02), but had a low probability of publication bias as reflected by the suggestive symmetry of the funnel plot (Figure 3a) and by Egger's test (P=0.45). Contrastingly, a comparison of allele ɛ4 with ɛ3 yielded a significant 81% increased risk for hypertension (95% CI: 1.41–2.32; P<0.0005; Figure 2b). However, significant between-study heterogeneity (I2=44.2%; P=0.02; Figure 3b) and publication bias (Egger's test: P=0.002) obscured the strength of this association. Given that, all but two study populations (one of Turks and one of Brazilians) were of Asian descent; we repeated the assessment of this association in Asian-descent populations and observed a slightly higher risk (OR=1.87; 95% CI: 1.41–2.32; P<0.0005; Supplementary Figure S1A). We found that heterogeneity was somewhat alleviated, and there was no apparent publication bias (Egger's test: P=0.108; Supplementary Figure S1B).
Forest plots (random effects model) of the hypertension risk associated with the ApoE gene ɛ2/ɛ3/ɛ4 alleles for ɛ2 vs. ɛ3 (a) and for ɛ4 vs. ɛ3 (b) in all the study populations. The summary odds ratio (OR) estimate is shown by the center of a solid diamond of which the left and right extremes represent the corresponding 95% confidence interval (95% CI). The area of the diamond is proportional to the weight of the study. The hollow diamond (and broken line) represents the overall risk estimate. The solid vertical line is set at the null value (OR=1.0). A full color version of this figure is available at the Hypertension Research journal online.
Funnel plots of the studies on the effect of the ɛ2/ɛ3/ɛ4 ApoE gene polymorphisms on hypertension risk for allele ɛ2 vs. ɛ3 contrast (a) and for allele ɛ4 vs. ɛ3 contrast (b). The vertical axis represents the log of the odds ratio (OR); the horizontal axis represents the s.e. of log (OR). The funnel plots are drawn with the 95% confidence limits. The symbols representing the data in the plot are sized proportionally to the inverse variance.
Association of the ApoE gene alleles with circulating cholesterols
Because seven studies, all of the subjects recruited from China, measured total cholesterol and high- and low-density lipoprotein cholesterol levels across the ɛ2 (ɛ2/2+ɛ2/3), ɛ3 (ɛ3/3) and ɛ4 (ɛ3/4+ɛ4/4) alleles, we performed an overall allele–phenotype association. Compared with subjects with the ɛ3 allele, those with the ɛ2 allele had significantly lower levels of total cholesterol (SMD=−0.69; 95% CI: −0.99 to −0.39; P<0.0005; Figure 4a) and low-density lipoprotein cholesterol (SMD=−0.44; 95% CI: −0.59 to −0.29; P<0.0005; Figure 5a), but had increased, albeit non-significant, high-density lipoprotein cholesterol levels (SMD=0.08; 95% CI: −0.06 to 0.21; P=0.258; Supplementary Figure S2A). By contrast, compared with subjects with the ɛ3 allele, those with the ɛ4 allele had significantly higher levels of total cholesterol (SMD=0.39; 95% CI: 0.2–0.57; P<0.0005; Figure 4b) and low-density lipoprotein cholesterol (SMD=0.43; 95% CI: 0.23–0.64; P<0.0005; Figure 5b), but no association was observed between the ɛ4 allele and high-density lipoprotein cholesterol levels (SMD=0.02; 95% CI: −0.16 to 0.19; P=0.852; Supplementary Figure S2B).
Forest plots (random effects model) of circulating cholesterol levels associated with the ApoE gene ɛ2/ɛ3/ɛ4 alleles for ɛ2 vs. ɛ3 (a) and for ɛ4 vs. ɛ3 (b) from the available studies. The vertical axis represents the standardized mean difference (SMD) of the circulating cholesterol levels. Each box represents the SMD estimate (a solid diamond), and the left and right extremes represent the corresponding 95% confidence interval (95% CI). The area is proportional to the weight of the study. The hollow diamond (and broken line) represents the overall SMD estimate. The solid vertical line is set at the null value (SMD=0). A full color version of this figure is available at the Hypertension Research journal online.
Forest plots (random effects model) of circulating low-density lipoprotein cholesterol levels associated with the ApoE gene ɛ2/ɛ3/ɛ4 alleles for ɛ2 vs. ɛ3 (a) and for ɛ4 vs. ɛ3 (b) from the available studies. The vertical axis represents the standardized mean difference (SMD) of circulating low-density lipoprotein cholesterol levels. Each box represents the SMD estimate (a solid diamond), and the left and right extremes represent the corresponding 95% confidence interval (95% CI). The area is proportional to the weight of the study. The hollow diamond (and broken line) represents the overall SMD estimate. The solid vertical line is set at the null value (SMD=0). A full color version of this figure is available at the Hypertension Research journal online.
Predicted association of circulating cholesterols with hypertension from Mendelian randomization in the mixed populations
Under the assumption of a linear-logistic relationship between circulating cholesterol levels and the odds of hypertension, statistical significance was reached only for total cholesterol and low-density lipoprotein cholesterol. Specifically, in comparing allele ɛ4 with ɛ3, the predicted OR for a 1 mmol l−1 increase in total cholesterol was 4.58 (95% CI: 1.83–67.21) when all qualified studies were included and 4.98 (95% CI: 1.94–76.36) in mixed populations of only Asian descent. With regard to low-density lipoprotein cholesterol, the predicted OR for a 1-mmol l−1 increase between allele ɛ4 vs. ɛ3 was 3.97 (95% CI: 1.71–43.48) in all populations and 4.29 (95% CI: 1.81–43.38) in Asian-descent populations. Because the 95% CIs of the predicted estimates above excluded the null hypothesis value of 1, we could safely reject the null hypothesis at the 5% level. Therefore, our data provided strong evidence for a causal influence of high circulating total cholesterol and low-density lipoprotein cholesterol levels on hypertension. Moreover, the direction of these predicted estimates was consistent with the association of the ApoE gene ɛ2/ɛ3/ɛ4 polymorphism with hypertension risk and circulating cholesterol levels.
Discussion
Using a comprehensive meta-analysis, we determined that the ApoE gene polymorphism ɛ2/ɛ3/ɛ4 is significantly associated with hypertension, which is in agreement with the results from two recent meta-analyses,13, 14 and is associated with large changes in circulating cholesterol levels. Under the rationale of the Mendelian randomization, we utilized this polymorphism as an instrumental variable, which is associated with circulating cholesterol levels, and we demonstrated the causal influence of high total cholesterol and low-density lipoprotein cholesterol levels on hypertension. To our knowledge, this is the first study to report the causality between high circulating cholesterol levels and hypertension, using the Mendelian randomization approach.
Hypertension has been strongly linked to a variety of lipoprotein metabolic abnormalities.41 The selection of the ɛ2/ɛ3/ɛ4 ApoE gene polymorphism as a tool to decipher cholesterol–hypertension causality is strongly supported by biological data, because it has been estimated that this polymorphism accounts for up to 11% of the total variance in circulating cholesterol levels in apparently healthy Caucasians.42 This is consistent with our pooled results, which indicated that a 1-mmol l−1 increase in total cholesterol corresponded to a 4.58-times higher likelihood of developing hypertension, and the same increase in low-density lipoprotein cholesterol corresponded to a 3.97-times higher likelihood. Because genotypes are invariant over time and can exert an effect on circulating cholesterol levels over a lifetime, the convincing ORs that related circulating total cholesterol and low-density lipoprotein cholesterol levels to hypertension risk in our Mendelian randomization analysis could represent the average life-time risk attributable to these cholesterols.
The differences in the bioavailability of the ɛ2/ɛ3/ɛ4 polymorphism proteins could be attributed to the fact that, because of the lack of a supradyl group, the under-oxidized ɛ4 lipoproteins are more easily cleared by scavenger receptors than are ɛ3 lipoproteins.43 Furthermore, according to the functional explorations, structural defects in the ApoE gene product might result in an impaired interaction between the ApoE-containing lipoproteins with their receptors and thus, induce the development of atherogenic dyslipidemias and premature cardiovascular diseases.44 ApoE gene knockout mice exhibited hypertension and endothelial dysfunction.45 Additionally, an ApoE gene polymorphism was found to significantly affect physiological activity by determining high-density lipoprotein cholesterol levels, indicating that environmental factors are important modulators of the effect of this polymorphism on circulating lipid levels.46 Therefore, one of this study's concerns, which has been regarded as a potential limitation of the Mendelian randomization, is related to the possible existence of pleiotropic effects of the ɛ2/ɛ3/ɛ4 polymorphism on other lipid or metabolic pathways that influence hypertension risk. However, we accepted the risk of introducing an error in the causal effect inferred from the instrumental variable analysis. Because very few studies have provided additional information concerning the distribution of other hypertension risk factors (such as body mass index, smoking and drinking) across the ApoE ɛ2/ɛ3/ɛ4 alleles, we cannot address the question of pleiotropy in the present study. If the data were available, we expect that they would show that the ɛ2/ɛ3/ɛ4 polymorphism is related to other risk factors.
We cannot exclude the possible linkage disequilibrium of the ɛ2/ɛ3/ɛ4 ApoE gene polymorphism with another genetic variant that is actually responsible for lipid variation, but there is no evidence that this is the case. However, as suggested by Sheehan et al.,5 it is somewhat irrelevant whether ɛ2/ɛ3/ɛ4 or another polymorphism is the causal variant for the phenotype when the two polymorphisms are in linkage disequilibrium, because either one could be considered an instrumental variable.
Other possible limitations to the use of the Mendelian randomization here include the possibility of population stratification, power deficiency, canalization, a lack of knowledge about the confounding factors and an inability to detect the effect of acute changes in the circulating cholesterol levels on hypertension.47 However, given the large sample sizes (n=5797) and the lack of publication bias in ethnically homogeneous subgroups, population stratification and power deficiency are unlikely to have a considerable impact on the results. Because canalization—a developmental compensation that can compensate for disruptive environmental or genetic forces5—is difficult to test outside of animal models, we cannot determine its effect in this meta-analysis. Although the effects of acute changes in circulating cholesterol levels on hypertension are not our major concern, several large-scale cohort studies have consistently reported the independent predictive value of some lipid fractions, such as total cholesterol, on hypertension risk in both adolescents and adults,1, 2, 3 indicating a smaller effect of acute cholesterol changes.
Even though our funnel plots and statistical tests showed low probability of publication bias, it remains a possibility, because we screened out the ‘gray literature’ (articles published in languages other than English and Chinese). However, the influence of publication bias on gene–disease association is expected to result in overestimation, not underestimation.48 Although publication bias might not affect the conclusions of this study, more studies are warranted to quantify the effect given the wide CIs obtained for the risk estimates. Moreover, with respect to the early onset of hypertension in youth, because most of the studies in this meta-analysis recruited subjects aged ⩾45 years (for whom environmental factors are likely to contribute more prominently than a genetic component to the development of hypertension),49 large association studies in a younger population of hypertensive subjects will be of added interest.
Despite the aforementioned limitations inherent in the Mendelian randomization, our results provide convincing evidence regarding the casual influence of high total cholesterol and low-density lipoprotein cholesterol levels on the development of hypertension, adding weight to the results from previous large-scale cohort studies. Although further analyses are warranted to evaluate the clinical utility of total cholesterol and low-density lipoprotein cholesterol levels in the treatment and control of hypertension, future studies should concentrate on elucidating the exact mechanisms by which these cholesterol levels can predict future hypertension.
References
Verdecchia P, Reboldi G, Angeli F, Gattobigio R, Borgioni C, Filippucci L, Poeta F, Porcellati C . Prognostic value of lipoprotein fractions in essential hypertension. Blood Press 2004; 13: 295–303.
Sesso HD, Buring JE, Chown MJ, Ridker PM, Gaziano JM . A prospective study of plasma lipid levels and hypertension in women. Arch Intern Med 2005; 165: 2420–2427.
Halperin RO, Sesso HD, Ma J, Buring JE, Stampfer MJ, Gaziano JM . Dyslipidemia and the risk of incident hypertension in men. Hypertension 2006; 47: 45–50.
Smith GD, Ebrahim S . Mendelian randomization: prospects, potentials, and limitations. Int J Epidemiol 2004; 33: 30–42.
Sheehan NA, Didelez V, Burton PR, Tobin MD . Mendelian randomisation and causal inference in observational epidemiology. PLoS Med 2008; 5: e177.
Ding EL, Song Y, Manson JE, Hunter DJ, Lee CC, Rifai N, Buring JE, Gaziano JM, Liu S . Sex hormone-binding globulin and risk of type 2 diabetes in women and men. N Engl J Med 2009; 361: 1152–1163.
Thomas DC, Conti DV . Commentary: the concept of ‘Mendelian Randomization’. Int J Epidemiol 2004; 33: 21–25.
Sorlí JV, Corella D, Francés F, Ramírez JB, González JI, Guillén M, Portolés O . The effect of the APOE polymorphism on HDL-C concentrations depends on the cholesterol ester transfer protein gene variation in a Southern European population. Clin Chim Acta 2006; 366: 196–203.
Maluf DG, Mas VR, Archer KJ, Yanek K, King A, Ferreira-Gonzalez A, Fisher RA, Posner M . Apolipoprotein E genotypes as predictors of high-risk groups for developing hyperlipidemia in kidney transplant recipients undergoing sirolimus treatment. Transplantation 2005; 80: 1705–1711.
Verghese PB, Castellano JM, Holtzman DM . Apolipoprotein E in Alzheimer's disease and other neurological disorders. Lancet Neurol 2011; 10: 241–252.
Abboud S, Viiri LE, Lütjohann D, Goebeler S, Luoto T, Friedrichs S, Desfontaines P, Gazagnes MD, Laloux P, Peeters A, Seeldrayers P, Lehtimaki T, Karhunen P, Pandolfo M, Laaksonen R . Associations of apolipoprotein E gene with ischemic stroke and intracranial atherosclerosis. Eur J Hum Genet 2008; 16: 955–960.
Ruixing Y, Jinzhen W, Weixiong L, Yuming C, Dezhai Y, Shangling P . The environmental and genetic evidence for the association of hyperlipidemia and hypertension. J Hypertens 2009; 27: 251–258.
Niu W, Qi Y, Qian Y, Gao P, Zhu D . The relationship between apolipoprotein E epsilon2/epsilon3/epsilon4 polymorphisms and hypertension: a meta-analysis of six studies comprising 1812 cases and 1762 controls. Hypertens Res 2009; 32: 1060–1066.
Niu WQ, Qi Y . Meta-based evidence for apolipoprotein E ɛ2/ɛ3/ɛ4 polymorphism in association with hypertension among Chinese. J Hum Hypertens 2011 (Epub ahead of print).
Shiwaku K, Gao TQ, Hojo N, Fukushima T, Yamane Y . Low levels of serum cholesterol and systolic blood pressure in Japanese with the apolipoprotein E3/2 genotype. Clin Chim Acta 1999; 284: 15–23.
Rastas S, Mattila K, Verkkoniemi A, Niinistö L, Juva K, Sulkava R, Länsimies E . Association of apolipoprotein E genotypes, blood pressure, blood lipids and ECG abnormalities in a general population aged 85+. BMC Geriatr 2004; 4: 1.
Niu W, Guo X, Su Y, Qiu C . Apolipoprotein E and low-density lipoprotein receptor gene polymorphisms in dyslipidemias-associated essential hypertension. J Hum Hypertens 2007; 21: 337–339.
Egert S, Boesch-Saadatmandi C, Wolffram S, Rimbach G, Müller MJ . Serum lipid and blood pressure responses to quercetin vary in overweight patients by apolipoprotein E genotype. J Nutr 2010; 140: 278–284.
Cohn LD, Becker BJ . How meta-analysis increases statistical power. Psychol Methods 2003; 8: 243–253.
Higgins JP, Thompson SG . Quantifying heterogeneity in a meta-analysis. Stat Med 2002; 21: 1539–1558.
Higgins JP, Thompson SG, Deeks JJ, Altman DG . Measuring inconsistency in meta-analyses. BMJ 2003; 327: 557–560.
Little J, Khoury MJ . Mendelian randomisation: a new spin or real progress? Lancet 2003; 362: 930–931.
Davey Smith G, Ebrahim S . Mendelian randomization’: can genetic epidemiology contribute to understanding environmental determinants of disease? Int J Epidemiol 2003; 32: 1–22.
Imazu M, Yamamoto H, Toyofuku M, Watanabe T, Okubo M, Egusa G, Yamakido M, Kohno N . Association of apolipoprotein E phenotype with hypertension in Japanese-Americans: data from the Hawaii-Los Angeles-Hiroshima Study. Hypertens Res 2001; 24: 523–529.
Yilmaz H, Isbir T, A∂açhan B, Aydin M . Is epsilon4 allele of apolipoprotein E associated with more severe end-organ damage in essential hypertension? Cell Biochem Funct 2001; 19: 191–195.
Li X, Du Y, Du Y, Huang X . Association of apolipoprotein E gene polymorphism with essential hypertension and its complications. Clin Exp Med 2003; 2: 175–179.
Bhavani AB, Sastry KB, Reddy NK, Padma T . Lipid profile and apolipoprotein E polymorphism in essential hypertension. Indian Heart J 2005; 57: 151–157.
Fuzikawa AK, Peixoto SV, Taufer M, Moriguchi EH, Lima-Costa MF . Association of ApoE polymorphisms with prevalent hypertension in 1406 older adults: the Bambuí Health Aging Study (BHAS). Braz J Med Biol Res 2008; 41: 89–94.
Hamulati W, Aisikaer Y, Yilihamujiang S . Relationship between ApoE gene polymorphism and patients of hypertension in Uighur. Chin J Immunol 2003; 19: 108–110.
Zhimian Z, Haiqing G, Qing S, Lihua F . Clinical study of relationship between ApoE gene polymorphism and hypertension in aged. Acta Academiae Medicinae Shandong 2000; 38: 173–176.
Caiyou H, Chenguang Z, Zeping L . The study of relation between blood pressure and apolipoprotein E genotypes in long-lived elderly in Bama area of Guangxi. Chin J Geriatr Heart Brain Vessel Dis 2007; 9: 11–13.
Weize Z, Xinghui L, Yongqing C . Relationship between apolipoprotein E gene polymorphism and essential hypertension in Han, Tibetan and Hui population in high altitude area. Chin Circ J 2007; 22: 263–266.
Chunli R, Yingxin P, Meixia L, Ruyi L, Xue G . Analysis of apolipoprotein E gene polymorphism in hypertensive patients. Shandong Med J 2007; 47: 29–30.
Yingwang L, Shuiping Z, An Z, Sai N . Effect of apolipoprotein E gene polymorphism on lipid and essential hypertension. Chin J Arterioscler 2002; 10: 517–520.
Wen P, Siming G, Wei L, Xiaohong Z . Apolipoprotein E gene polymorphism and hypertension. J Chin Gen Pract 2003; 2: 14–15.
Guofang C, Zhu M, Bingxian H . Relationships between polymorphism of apoE gene and hypertension in the Han ethnics of China. Chin J Clin Rehab 2003; 7: 2545–2546.
Min P, Jianhua Z, Li L . Association of Apolipoprotein E with essential hypertension, NIDDM and coronary heart disease in Chinese population. Chin Synth Med 2002; 3: 1–3.
Huaiqin Z, Weijian YKH, Jie L, Chengang S . To explore the relativity of apolipoprotein B and E gene polymorphism with coronary heart disease and hypertension. Prev Treat Cardio-Cerebral-Vasc Dis 2002; 2: 20–22.
Haiqing G, Junli Y, Zhimian Z, Qing S, Huanqin C, Yongle S, Guihua W . Clinical study on apolipoprotein E genetic polymorphism among elders. Chin J Geriatr 2002; 21: 299–300.
Kesäniemi YA, Ehnholm C, Miettinen TA . Intestinal cholesterol absorption efficiency in man is related to apoprotein E phenotype. J Clin Invest 1987; 80: 578–581.
Ciolac EG, Bocchi EA, Bortolotto LA, Carvalho VO, Greve JM, Guimarães GV . Haemodynamic, metabolic and neuro-humoral abnormalities in young normotensive women at high familial risk for hypertension. J Hum Hypertens 2010; 24: 814–822.
Davignon J, Gregg RE, Sing CF . Apolipoprotein E polymorphism and atherosclerosis. Arteriosclerosis 1988; 8: 1–21.
Humphries SE, Talmud PJ, Hawe E, Bolla M, Day IN, Miller GJ . Apolipoprotein E4 and coronary heart disease in middle-aged men who smoke: a prospective study. Lancet 2001; 358: 115–119.
Brewer Jr HB, Zech LA, Gregg RE, Schwartz D, Schaefer EJ . Type III hyperlipoproteinemia: diagnosis, molecular defects, pathology, and treatment. Ann Intern Med 1983; 98: 623–640.
Yang R, Powell-Braxton L, Ogaoawara AK, Dybdal N, Bunting S, Ohneda O, Jin H . Hypertension and endothelial dysfunction in apolipoprotein E knockout mice. Arterioscler Thromb Vasc Biol 1999; 19: 2762–2768.
Corella D, Guillén M, Sáiz C, Portolés O, Sabater A, Cortina S, Folch J, González JI, Ordovas JM . Environmental factors modulate the effect of the APOE genetic polymorphism on plasma lipid concentrations: ecogenetic studies in a Mediterranean Spanish population. Metabolism 2001; 50: 936–944.
Timpson NJ, Harbord R, Davey Smith G, Zacho J, Tybjaerg-Hansen A, Nordestgaard BG . Does greater adiposity increase blood pressure and hypertension risk?: Mendelian randomization using the FTO/MC4R genotype. Hypertension 2009; 54: 84–90.
Smith GD, Harbord R, Milton J, Ebrahim S, Sterne JA . Does elevated plasma fibrinogen increase the risk of coronary heart disease? Evidence from a meta-analysis of genetic association studies. Arterioscler Thromb Vasc Biol 2005; 25: 2228–2233.
Pereira TV, Nunes AC, Rudnicki M, Yamada Y, Pereira AC, Krieger JE . Meta-analysis of the association of 4 angiotensinogen polymorphisms with essential hypertension: a role beyond M235T? Hypertension 2008; 51: 778–783.
Acknowledgements
This work was supported by the SMC–Excellent Young Faculty Award of Shanghai Jiao Tong University, the Youth Foundation of Shanghai Municipal Health Bureau (2010Y050), the Shanghai Rising Star Program (11QA1405500), and the National Science Foundation for Young Scientists of China (30900808, 81000109).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies the paper on Hypertension Research website
Rights and permissions
About this article
Cite this article
Niu, W., Zhang, X. & Qi, Y. Association of an apolipoprotein E polymorphism with circulating cholesterols and hypertension: a meta-based Mendelian randomization analysis. Hypertens Res 35, 434–440 (2012). https://doi.org/10.1038/hr.2011.202
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/hr.2011.202
Keywords
This article is cited by
-
Causal association of circulating cholesterol levels with dementia: a mendelian randomization meta-analysis
Translational Psychiatry (2020)
-
Causal relevance of circulating high-density lipoprotein cholesterol with cancer: a Mendelian randomization meta-analysis
Scientific Reports (2015)
-
Studies of the CETP TaqIB and ApoE Polymorphisms in Southern Thai Subjects with the Metabolic Syndrome
Biochemical Genetics (2015)