Abstract
For any given genotype, the environment in which an apple is grown can influence the properties of the fruit considerably. While there has been extensive research on the mechanism of the genetic control of fruit quality traits, less effort has been made to investigate the way that these genetic mechanisms interact with the environment. To address this issue, we employed a large ‘Royal Gala’ × ‘Braeburn’ population of 572 seedlings replicated over sites in three climatically diverse apple-growing regions in New Zealand. Phenotyping for traits including fruit maturation timing, firmness and dry matter content was performed at each of these three sites for a single growing season (2011), and at two sites (Motueka and Hawke's Bay) for two seasons (2009 and 2010). The phenotype data collected over 2 years at two sites enabled the detection of 190 quantitative trait loci (QTL) that controlled these traits regardless of year or growing location, as well as some chromosomal loci that influenced the traits in a single given environment or year. For those loci that were environmentally stable over three sites, there was an interdependency of fruit maturation date, dry matter content and storage potential within this population, with two regions on Linkage Groups (LGs) 10 and 16 strongly contributing. If these loci were used in a marker-assisted selection programme to select for progeny bearing firmer fruit, this would have the unintentional consequence of selecting, high dry matter content, later maturing apples. In addition, a further 113 new QTLs with a smaller effect were identified, some of which were exhibited only in a single growing environment, demonstrating the underlying complexity of control of traits determining fruit quality, in addition to the need for being aware of environmental effects when developing new apple varieties.
Similar content being viewed by others
Introduction
In clonally propagated horticultural crops, adaptation to different growing environments can lead to considerable differences in fruit quality. Anecdotally, it is well known by fruit growers that some years or growing seasons result in better quality fruit, and some areas are not suited for some crops. For example, there are numerous examples of reports of the effect on growing environment on grapes where ‘terroir’ influences wine characters.1 In apples, it has been shown that the reddening of the skin is controlled by environmental temperature2 and the changing environment has been reported recently to be altering the quality of apple fruit.3 Most of these trait-focused studies have used commercially grown horticultural crops grown in different regions and there are few reports to date on the interaction of trait genetic control mechanisms with the growing environment.
In apple, texture is a key quality determinant for consumers.4 Although fruit maturation and ripening are key developmental processes that underlie firmness and texture, there is currently a limited understanding of the controlling mechanisms. Current models point to both developmental and environmental control of ripening,5 mediated often through the ripening hormone ethylene, suppression of which reduces ripening in many fruits.6,7 It has been shown that within a genotype, there is an association between dry matter (DM) content, fruit firmness and storage.8,9 Apart from texture, DM is also an important determinant for other aspects of fruit quality related to taste (sugar and acid metabolism), where consumers have shown a preference for fruit with higher DM.8 Apple loss of firmness during storage is strongly controlled by ethylene and its suppression causes the fruit to remain firmer.10 However, genotypic differences in ethylene biosynthesis do not solely account for differences in ripening rates,11 suggesting that differences in ethylene perception and downstream signalling are also critical. There is a suite of cell wall-associated enzymes that change in expression12–14 during ripening and quantitative trait loci (QTL) analysis has linked apple fruit softening with genetic loci that contain the ethylene biosynthesis genes 1-AMINO-CYCLOPROPANE-CARBOXYLASE SYNTHASE 1 and 3 (ACS1, ACS3),11,15 and 1-AMINO-CYCLOPROPANE-CARBOXYLASE OXIDASE 1 (ACO1),16 cell wall loosening genes (EXPANSIN – EXP7)17 and POLYGALACTURONASE 1 (PG1),18 as well as cell wall genes such as PECTIN LYASE (Md-Pel) and XYLOGLUCAN ENDOTRANSGLYCOSILASE (Md-XET).19 This is supported by molecular studies showing that at the gene expression level, there is a tight correlation between the cell wall gene PG1 and loss of firmness during storage14 and when PG1 is suppressed, apples remain firmer for longer in storage.20
In this study, a segregating population was used to assess the contribution of genetic makeup and growing environment on fruit maturation and fruit quality. Two contemporary commercial apple cultivars, ‘Royal Gala’ and ‘Braeburn’, were chosen as parents, as breeding populations derived from these individuals have produced new cultivars in which fruit firmness traits exceed those of either of the parents.21 This mapping population was replicated in different geographic locations to identify genomic loci whose expression not only is important for regulating maturation and ripening, but also is influenced by growing environment.
Materials and methods
Plant material and fruit sampling
A segregating F1 population of 599 seedlings from a ‘Royal Gala’ × ‘Braeburn’ cross was planted out in a split-plot layout across three diverse geographic locations, one in each of three apple production regions in New Zealand, to investigate the Genotype by Environmental (GxE) control of time to harvest, DM content and fruit firmness. The 599 seedlings were each replicated by grafting onto six ‘M27’ dwarfing rootstocks and planted in duplicate in each of the Plant & Food Research Havelock North, Motueka and Clyde research orchards (Havelock North, Hawke's Bay (lat. 39°40′S, long. 176°53′E), Riwaka, Motueka (lat. 41°6′S, long. 172°58′E) and Clyde, Otago (lat. 45°12′S, long. 169°19′E)). Trees were planted 3×1.5 m apart in an east-west orientation with ‘genotype’ the main effect (n=6) and ‘environment’ the subplot (n=2). The ‘Royal Gala’ and ‘Braeburn’ parents grafted onto ‘M9’ rootstocks were planted one after every 10 genotypes, alternating, along the rows within the same blocks (Supplementary Fig. S1). This parental pattern enabled confirmation that there was no within-plot variability that compromised expression of fruit traits. Pests and diseases were controlled by conventional orchard techniques according to standard commercial practices. For each growing season the trees were treated as consistently as possible, with a standardized low crop load and supplementary irrigation when conditions required. In the first year in the orchard (2006), fruit were removed shortly after fertilisation to promote strong vegetative growth. In subsequent seasons, the apples were thinned to a low crop load (10 fruit per tree in the second year, 20–25 fruit per tree in the third year, then 6 fruit per cm2 trunk cross-sectional area thereafter).
Phenotyping
To assess the orchard variation at any given site, 15–40 fruit from each of the ‘Royal Gala’ and ‘Braeburn’ parents were assessed for each of the traits in two orchards (Hawke's Bay and Motueka). The first harvest assessments were made in 2009 at Hawke's Bay and Motueka and the second in 2010 at the same two sites. A third harvest was performed at all three sites in 2011 using a subset of 127 seedlings selected for carrying high firmness alleles of QTLs linked to PG1 and ACO1. Fruit were harvested at a standardized maturity, based on background colour change and starch clearance (1.5–2.5 units on a New Zealand industry generic 0–6 Starch Pattern Index scale). Ten representative fruit per tree were harvested and for five of these fruit, fresh weight was recorded and maximum firmness measured using a GUSS penetrometer with an 11.1-mm probe inserted at a speed of 10 mm per second. Soluble solids content was measured using an Atago PAL-1 pocket digital refractometer, Starch Pattern Index assessed and DM measured as described by Palmer et al.22 The remaining five fruits were stored in cartons lined with perforated polythene bags to minimize water loss, at 0.5 °C for 10 weeks and at 20 °C for 7 days, before assessment for fruit firmness and soluble solids content.
DNA extraction and single nucleotide polymorphism (SNP) marker development
Genomic DNA was extracted from young leaves from the parents and the 599 seedlings of the ‘Royal Gala’ × ‘Braeburn’ segregating population using the Qiagen Plant DNeasy Plant Mini kit (Qiagen, Hilden, Germany) following the manufacturer's protocol. Ten nanograms of genomic DNA from each sample were then amplified by whole-genome amplification23 using the GenomiPhi V2 DNA Amplification Kit (GE Healthcare, Little Chalfont, Buckinghamshire, UK). A subset of the 951 SNP markers used for anchoring the ‘Golden Delicious’ genome sequence to a genetic map was selected for map construction, on the basis of even distribution along the ‘Golden Delicious’ pseudo-chromosomes. SNPlex (Applied Biosystems Inc., Foster City, CA, USA) genotyping assays were carried out using 1 µL (from 45 to 225 ng) of WGA-DNA according to the manufacturer's protocol. Samples were run on a 3730xl DNA Analyzer (Applied Biosystems Inc.) and data were analysed using the Gene Mapper v.4.0 software (Applied Biosystems Inc.). Genotype analysis was performed according to the SNPlex_Rules_3730 method, in accordance with the manufacturer's default settings.
Genetic map construction and QTL analysis
Seventeen individuals were removed from the population as the SNP marker data indicated they were potential pollen contaminants, resulting in a set of 572 true seedlings usable for map construction. The genetic maps used for QTL analysis were the parental maps of the ‘Royal Gala’ × ‘Braeburn’ population as described in Chagné et al.24 In brief, a subset of 132 SNP markers was used to construct a ‘Braeburn’ genetic map with an even marker distribution, favouring backcross type markers (ab × aa) above less informative intercross markers (ab × ab). This map spanned a cumulative distance of 1004.8 cM, covered all 17 linkage groups (LGs) and had an average of one marker every 7.6 cM. One linkage group (LG 7) spanned only 4.1 cM and had only three markers. A further subset of 132 SNP markers was used to build the ‘Royal Gala’ genetic map, using similar distribution and segregation type criteria as for the ‘Braeburn’ map. The cumulative genetic length of the ‘Royal Gala’ genetic map was 863.9 cM, covering all 17 LGs, with an average of one marker every 6.7 cM. Genetic markers for candidate genes related to fruit texture were then located on the map (Supplementary Table S1). These included genetic markers for Polygalacturonase (PG1) and AAC oxidase (ACO1) on LG10, a duplicated copy of PG on LG5 and ethylene-related genes identified using a microarray experiment.25
QTL analysis was performed with MapQTL version 5.0. For the interval mapping analysis, the LOD threshold for significance of a QTL was calculated at the genome level using 1,000 permutations. Only the QTLs with a LOD score significant at greater than 90% genome-wide were retained. The most significant marker for each QTL was then used as a cofactor for a multiple QTL analysis (MQM) for detecting minor QTLs that were concealed by the major QTL in the previous interval mapping analysis. The percentage of phenotypic variation explained by all detected QTLs for one trait was calculated using a linear regression fitting the QTLs detected for each trait as factors.
Results
Growth and fruit assessment of a ‘Royal Gala’ × ‘Braeburn’ seedling population
The three apple-growing regions in New Zealand where the replicated 599 ‘Royal Gala’ × ‘Braeburn’ mapping population seedlings were grown differed from one another in key environmental factors affecting growth, such as temperature and rainfall (Figure 1), sunshine hours, growing degree days and soil types. Havelock North (Hawke's Bay) and Motueka (Nelson region) are coastal maritime environments, while Clyde (Central Otago) is an inter-montane semi-arid basin. In particular, rainfall is higher in Hawke's Bay than in Motueka in January, while it is higher in Motueka than Hawke's Bay from September to December; however, the temperature is similar in Hawke's Bay and Motueka during apple fruit growth and ripening.
Environmental variation between apple-growing sites in New Zealand. Average rainfall (histograms) and temperature (curves) were calculated for the three growing seasons used for the phenotyping of the ‘Royal Gala’ × ‘Braeburn’ segregating population (2009–2011). The location of the three sites is indicated on the map of New Zealand (top left). The periods of fruit development and apple ripening are indicated.
For the first year (2009), when both tree replicates of fruit grown and harvested at standard maturity in Hawke's Bay and Motueka were assessed, fruit maturation date ranged from 27 January to 30 April (Table 1). There was little variation between tree replicates for both DM and fruit firmness within a genotype (seedling) at each orchard site both for the samples assessed at maturity and those assessed following storage (Supplementary Table S2a), suggesting that phenotypic responses for the segregating population can be attributed to genetic effects rather than within-orchard effects. Because of this lack of within-orchard variation between genotypes and the lack of variation among fruit harvested from the same trees (Supplementary Table S2b), the replicated fruit from replicated individuals at each site were averaged for further analysis. For the following year (2010), only a single replicate tree was assessed in each of these sites. Within the seedling population, there was a significant effect of the genotype and site for all measured variables (p<0.001; Table 2). The year effect was significant (p<0.001) for all variables except firmness measured at harvest and loss of firmness during storage. The year×site effect was not significant for loss of firmness after storage and fruit maturation date. The genotype x year effect was not significant for any variable, indicating that the genetic effect was consistent across years. The correlations between traits across two sites in 2009 and 2010 are presented in Table 3. The year to year correlation between 2009 and 2010 for all traits ranged from r=0.31 to 0.88 (Table 3). The between-site correlation ranged between r=0.43 and 0.80. For each of the traits the correlations between years at the same site and between site during the same year were generally high (Table 3; p<0.01).
Fruit matured later in Motueka than in Hawke's Bay, by 9–12 days, consistently over two years of measurement. There was a strong genetic control of fruit maturation date, with a correlation between site and year, ranging from r=0.73 to 0.87 (Table 3). Fruit maturation date was significantly correlated to textural traits, with the highest correlation coefficient being r=−0.47 for loss of firmness in Motueka in 2010. Interestingly, the harvest date was less correlated to firmness measured at harvest than firmness measured after storage (Figure 2b). Harvest date was negatively correlated to loss of firmness, meaning a weak trend for early maturing genotypes to soften more rapidly during storage. For fruit firmness, the apples grown in Motueka tended to soften more than those in Hawke's Bay (Table 1); however, there was no trend for which site had firmer fruit at harvest and after storage between the two years of measurement (Figure 2a). DM content was found to be slightly higher in Hawke's Bay than in Motueka, especially in 2010 (Table 1). The 2011 data collected on three sites were not included in the variance and correlation analysis as the population was biased toward high firmness alleles at one locus.
Analysis of apple fruit firmness data between sites. (a) Distribution of firmness at harvest of the 572 genotypes grown at Hawke's Bay (HB—red) and Motueka (Mot—green), in 2009 (dark) and 2010 (light). (b) Distribution of firmness following storage of the 572 genotypes grown at Hawke's Bay (HB—red) and Motueka (Mot—green), in 2009 (dark bars) and 2010 (light bars). (c) Nine genotypes selected for extremes of environmental regulation. Fruit that had significantly different firmness between the two sites at harvest and following storage in two environments over 2 years (*p<0.01, **p<0.01).
Fruit firmness variability between sites
Variability between two sites and two years was observed in the ‘Royal Gala’ × ‘Braeburn’ population for fruit firmness at harvest and after storage (Figure 2a and 2b). From this dataset it was clear that there were genotypes whose firmness were strongly influenced by environment and those that were less so. In the 2010 data, there were genotypes that had significantly (p<0.01) higher firmness both at harvest and following storage in different sites (Figure 2c), as well as genotypes that appeared to be completely unaffected by the environment between these two orchards (Figure 2c, genotypes 209, 228 and 70). Interestingly, the firmness at harvest was not correlated with the firmness following storage at each of the sites, with some genotypes having similar firmness at harvest and considerably different firmness following storage (Figure 2c, genotypes 183 and 331).
Identification of QTLs for harvest time, firmness and DM content
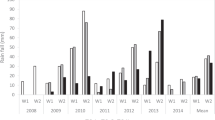
‘Royal Gala’ and ‘Braeburn’ parental genetic maps24 (Supplementary Fig. S2) constructed using SNP data and enriched using candidate gene-based markers (Supplementary Table S1) were employed to identify QTLs for harvest date, fruit firmness and DM. In total, 190 QTLs were identified independently for each of the traits phenotyped over two years (2009 and 2010) and two sites (Hawke's Bay and Motueka). These clustered into 35 unique genomic regions inherited from either parent. Thirty-two QTLs were detected for firmness at harvest, clustering into 14 unique genomic regions (Supplementary Table S3a), 45 QTLs for firmness after storage (Supplementary Table S3b), 32 QTLs for loss of firmness during storage clustering into 14 unique genomic regions (Supplementary Table S3c), 30 QTLs for DM content clustering into 16 unique genomic regions (Supplemental Table 3d), and 51 QTLs for harvest date clustering into 16 unique genomic regions (Supplementary Table S3e). The effects expressed as percentages of the phenotypic variation explained by each QTL independently are presented in Figure 3. In addition, 29 QTLs were detected in 2011 over all three sites using a subset of 127 seedlings of the population that was selected to carry the favourable QTL alleles of LG10 from both parents (PG1-BB, PG1-RG and ACO1-BB favourable allele was selected). Here the QTL effect explained from 3.2% to 52.9% of explained the phenotypic variation (Supplementary Table S3f). The percentage of the phenotypic variation explained by all QTLs for each trait in this study is presented in Table 4 and compared with the percentage variation calculated using the data for previously published QTLs26,27 on LG10 and LG16 only.
Percentage of the phenotypic variation explained by the QTLs detected in the ‘Royal Gala’ × ‘Braeburn’ apple population. The percentages of the variance explained were obtained for each QTL independently using MapQTL 5.0. QTL clusters are indicated as in Supplementary Table S3. QTLs shown in red were previously published,17,19,26–29 while QTLs in grey are new to this study.
Effect of environment on QTL stability and interaction among QTLs
Stable QTL regions for harvest date, firmness at harvest, firmness following storage, loss of firmness during storage, and DM are located on all LGs except LGs 4, 7, 12 and 17 (Figures 4 and 5 for the ‘Royal Gala’ and ‘Braeburn’ parents, respectively) with one unstable QTL detected for one trait, one site and one year on LG12. With QTLs located on 10 different LGs, harvest date is the trait with the most QTLs for control. Seventeen QTL clusters for fruit firmness coincided with QTLs previously detected in apple on LG1 (clusters EXP-BB and EXP-RG),17 LG3 (Lg3a-BB and Lg3b-BB),28 LG10 (ACO1-BB, PG1-BB, PG1-RG),26 LG14 (FUL-BB),29 LG15 (Lg15a-BB, Lg15a-RG)30 and LG16 (Lg16-BB).27 The QTLs from LGs 1, 10, 14, 15 and 16 explained the major variation for fruit firmness both at and after harvest and for loss of fruit firmness during storage in the population (Figure 3) and were stable between sites and between years. Two QTLs for fruit firmness after storage and loss of firmness during storage were detected on LG10 when using MQM analysis.
Detection of QTLs for control of fruit firmness, harvest date and dry matter content in ‘Royal Gala’ apple. LOD scores were plotted against the location on the ‘Royal Gala’ genetic map using the phenotypic data collected in Motueka (Mot—green dots) and Hawke's Bay (HB—red dots) in 2009 and 2010. The dashed line represents QTL significance at 95% genome-wide. The location of candidate genes is indicated using black arrows: EXPANSIN (Exp7), POLYGALACTURONASE (PG), ACC OXIDASE (ACO) and ACC SYNTHASE (ACS).
Detection of QTLs for control of fruit firmness, harvest date and dry matter content in ‘Braeburn’ apple. LOD scores were plotted against the location on the ‘Braeburn’ genetic map using the phenotypic data collected in Motueka (Mot—green dots) and Hawke's Bay (HB—red dots) in 2009 and 2010. The dashed line represents QTL significance at 95% genome-wide. The location of candidate genes is indicated using black arrows: EXPANSIN (Exp7), POLYGALACTURONASE (PG), ACC OXIDASE (ACO), FRUITFUL (FUL) and ACC SYNTHASE (ACS).
QTLs were present or absent in different environments (sites and years). Five QTL regions stable across 2 years were detected in fruit grown in Motueka but not in Hawke's Bay. These were located on LG6 (LG6a-BB and LG6a-RG) for control of fruit firmness at harvest, and LG5 (LG5a-RG and LG5a-BB) for change of fruit firmness during storage and LG9 for fruit firmness after storage (LG9a-RG), respectively, indicating the presence of site-specific QTLs.
QTLs also explained variable phenotypic variation between traits. For example, while the QTL on LG16 (LG16-BB) explained from 3.3% to 7.7% of the phenotypic variation for harvest fruit firmness, it had a larger effect on loss of fruit firmness during storage (8.2%–13.1%). This highlights the complex interaction of QTLs in controlling fruit firmness after storage. Fruit firmness after storage related to both fruit firmness at harvest, the rate of fruit softening and the interaction between them. The QTLs detected could then be separated into three groups: (i) QTLs influencing both fruit firmness at harvest and firmness change during storage on LGs 10 and 15; (ii) QTLs for which postharvest fruit firmness was due to harvest fruit firmness but not fruit softening on LGs 1, 6 and 14; and (iii) QTLs associated with softening but not fruit firmness at harvest on LGs 10, 5, 3 and 16. Stable QTLs for DM content colocated with QTLs for fruit firmness on LGs 1, 9 and 16.
Growth environment and genetic linkages between fruit maturation date and loss of firmness during storage
QTLs in common were identified between control of fruit maturation date and loss of firmness during storage, especially on LGs 10 (QTL clusters PG1-BB, ACO1-BB and PG1-RG) and 16 (Lg16-BB). Close inspection of the LOD score peak position indicates it is likely that these QTLs are pleiotropic, i.e., explaining both traits in a dependent manner, as the LOD score peaks for harvest date and loss of firmness coincided for QTLs on both LGs (Figures 4 and 5), so that seedlings carrying LG 10 and 16 QTL alleles for control of fruit firmness had early maturing fruit. However, the QTLs related to fruit firmness at harvest on LGs 1 and 14 were independent of harvest date. Interestingly, the LG10 and LG16 QTLs explained more of the phenotypic variation of fruit firmness at harvest in Hawke's Bay than in Motueka (Table 4); however, the trend was opposite for loss of firmness during storage and fruit maturation date, and the QTLs explained more of the phenotypic variation in Motueka than in Hawke's Bay.
Candidate genes associated with QTLs
The QTLs on LG10 that influenced fruit firmness colocated with ACO1 (QTL cluster ACO1-BB) and PG1 (PG1-RG and PG1-BB). QTLs controlling fruit firmness at harvest were identified in the LG1 region colocating with EXP7 (EXP-RG); however, the QTL detected for fruit firmness on LG15 (Lg15a-BB and Lg15a-RG) did not colocate with ACS1, as ACS1 maps to the bottom of LG15, whereas the peak of the QTL maps consistently at the top of the LG. A QTL for firmness after storage at the top of LG3 (Lg3a-RG and Lg3a-BB) colocated with candidate gene EB137463, which has homology to a BZIP transcription factor. The QTL on LG16 from the ‘Braeburn’ map (Lg16-BB) is of considerable interest as it controlled fruit maturation date, fruit firmness and DM content. The QTL was most significant with markers located between GDsnp00071 and GDsnp00555. Polymorphisms within this area were identified and six additional markers were mapped to increase the density in the region on the ‘Braeburn’ genetic map between 28.7 and 36.9 cM. Using the draft genome of ‘Golden Delicious’, a 2.4 Mb genomic region between these markers was extracted that contained 411 predicted gene models in the ‘Golden Delicious’ genome assembly; these can be seen in Supplementary Table S4.
Discussion
Complexity of control of apple fruit quality can only be demonstrated in a large multisite segregating population
The high number of QTLs (190) that were identified for the genetic control of aspects of fruit quality, including fruit firmness at and after harvest, DM content and harvest date in the ‘Royal Gala’ × ‘Braeburn’ segregating population, demonstrates the complexity of control of these attributes in apple fruit. Our comprehensive study identified 113 previously unreported QTLs, including 24 new clusters and QTLs that become apparent only when populations were grown in different environments. In addition, we confirmed previously reported large effect QTLs for fruit firmness.16,17,20,26–28,31–36 Our use of a large replicated segregating population of 572 seedlings phenotyped over 2 years was critical for the detection of small effect QTLs explaining less than 10% of the phenotypic variation and the previously unreported site-specific QTLs. Most of the small-effect QTLs were stable across years, with LOD scores that were significant at genome-wide level as well as chromosome-wide level. It is well known that the power of QTL detection for accurate estimation of the effect of QTLs and for identifying smaller-effect QTLs is highly dependent on the size of the segregating population, and to a lesser extent on the density of genetic markers.37 Most previous QTL studies performed in apple and other fruit tree species were undertaken at a single geographic location, using segregating populations of fewer than 300 seedlings. In contrast, our study was carried out across three sites, three seasons and almost doubled the population size of previous studies. This enhanced significantly the power of QTL analysis for fruit traits, demonstrated by the increase of number of QTLs detected for previously reported traits. The only other reported biparental population and multisite apple QTL mapping experiment was carried out using 251 seedlings from a ‘Fiesta’ × ‘Discovery’ population replicated in three locations in Switzerland.28 Here, no differences were identified among the three sites for QTLs for fruit firmness at harvest, indicating that the differences in environmental conditions among the three sites did not allow the detection of site-specific QTLs and also that the size of the segregating population may have not allowed the detection of site-specific and other small-effect QTLs. A recent study used Bayesian QTL analysis using a total of 1347 individuals from 27 interconnected populations of average size of 50 individuals grown in five sites30 and phenotyped for fruit firmness after storage across 3 years. Our study confirmed all detected QTLs from this large study, which indicates that biparental populations, when using sufficient number of seedlings and multiple replicates, are not underpowered compared with studies using multiple populations. Another recent example of how crucial it is to use a large segregating population for QTL analysis of complex traits, is the mapping of fruit firmness and softening using a ‘Fuji’ × ‘Mondial Gala’ population of 176 full-sib individuals.26 Although this study used a parent in common with our study (a sport of ‘Gala’) and similar phenotyping methods, QTLs were detected only on LG10 and the majority of these were below the threshold for genome-wide significance. Our larger population using the same parent enabled confirmation of the role of the large-effect QTLs on LG10 that colocate with PG1 and ACO1 and in addition, a further five QTLs for fruit firmness on LGs 1, 6, 9, 14 and 15 of ‘Gala’ were identified.
Of particular interest are the QTLs that were detected in a single environment only. For example, the LG6 and LG9 QTLs that contribute to higher fruit firmness at harvest and postharvest, with no influence on softening rate, were identified only in the population grown in Motueka (Supplementary Table S3). Furthermore, the LG10 and LG16 QTLs explained more of the phenotypic variation for fruit firmness at harvest in Hawke's Bay growing conditions than in Motueka, and more of the phenotypic variation for loss of firmness in Motueka than in Hawke's Bay. This may indicate that expression of the genes controlling the QTLs for loss of firmness was activated by the Motueka climatic or edaphic conditions. In particular, the stronger effect of the LG10 QTL (PG1-RG; Figure 3) in Motueka may confirm an effect of the role of low temperature on the activation of PG1.5 Nevertheless, Hawke's Bay and Motueka have similar average monthly temperatures (Figure 1). Further analysis of the expression and regulation of PG1 at a finer physiological and climatic scale is required to identify which conditions are responsible for the difference in fruit firmness between PG1 alleles. The strong environmental effects on some genotypes suggest that the seedling evaluation stage of breeding programs should be located in the regions for planned production. However, the identification of some seedling genotypes with consistent fruit quality over different sites suggests that next-generation apple cultivars could be selected for production over a range of environments, by phenotyping over a range of sites.
Genetic control of fruit dry matter content and its significance for apple breeding
Current apple breeding programmes have not selected for DM content, but for other related fruit characters such as fruit sensory attributes—flavour, firmness and crispness—which are all influenced by DM,9 as well as for tree characters such as pest and disease resistance. We report the first QTLs identified for apple fruit DM content, which are located on ten LGs and account variously for from 3% to 21.8% of the phenotypic variation. Apart from a small-effect QTL for DM content on LG4, all DM content QTLs colocated with fruit firmness QTLs, which supports observations linking good-storing apples with high DM in ‘Royal Gala’.9 Because of its simplicity in measurement, DM is becoming a common metric for fruit quality; however, the molecular control of DM content has not been explored fully in different fruit species. The DM content in kiwifruit is predominantly influenced by starch38 and in other species QTLs controlling fruit DM content have been reported for peach39 and tomato.40
Candidate genes for apple fruit firmness control
The ‘Royal Gala’ and ‘Braeburn’ parental genetic maps were constructed using SNP markers that are anchored to the apple genome assembly of ‘Golden Delicious’,41 which enables the identification of positional candidate genes underlying QTLs. These candidate genes were mapped using HRM-based markers to confirm the colocation with QTLs. The two largest and most stable QTLs were located on LG10, on different positions, and colocated with the PG1 and ACO1 candidate genes respectively (Figures 4 and 5), confirming QTLs identified previously.18,19,26,27,30,34,42 Both QTLs were associated with all measured traits; however, the percentage of phenotypic variation they explain is strongest for loss of firmness, which strengthens their key role in fruit softening. As both ‘Royal Gala’ and ‘Braeburn’ are heterozygous for the ACO1-1 and ACO1-2 alleles,43 it is not surprising to find QTLs for loss of fruit firmness colocating with ACO1. ACO1 colocation with QTLs for loss of firmness is consistent with its expression during fruit ripening in ‘Royal Gala’26,44 and ‘Braeburn’,44 and with colocation with loss of firmness in other studies in apple.16 Interestingly, we did not identify any colocation between firmness QTLs and ACS1 on LG15, although this candidate gene was linked to apple shelf life11,15,16 and to fruit ethylene production16 in other studies. PG1 colocation with loss of firmness QTLs is consistent with results obtained in other populations18,19,26,42 and with the fact that its downregulation affects fruit firmness.20 The year-stable LG16 QTL from ‘Braeburn’ confirms this QTL previously identified in the ‘Telamon’ × ‘Braeburn’,27 ‘Fuji’ × ‘Pink Lady’19 and HiDras30 populations, but we were unable to identify a candidate gene for this QTL, in spite of searching a set of 411 positional candidate genes identified in the ‘Golden Delicious’ genome. A putative BZIP transcription factor (EB137463) was found to colocate with a QTL for storage firmness and loss of firmness at the proximal end of LG3. Two stable QTLs on LG1 and LG14 were detected for firmness at harvest only, and not for loss of firmness. The LG14 QTL colocated with the apple homologue of FRUITFUL and a marker (MdMADS2.1) derived from this was associated with fruit firmness previously.29,30 The LG1 QTL colocated with an expansin (Exp7), which has been associated with fruit firmness at harvest and after storage, but not the softening rate.17
Relationship between fruit firmness and time of maturity
Empirical evidence indicates that early season apples tend to soften more during storage. This is the case with the parent ‘Royal Gala’, which is early season and has an average ability for storage in relation to mean loss of firmness values.45 In the ‘Royal Gala’ × ‘Braeburn’ progeny of 572 individuals, significant correlations were observed across years and sites between harvest date, which was determined at a consistent Starch Pattern Index value, and fruit firmness. Stronger correlation coefficients with harvest date were observed for firmness measured after storage and loss of firmness during storage, than for firmness measured at harvest (Table 3). The largest-effect QTLs for harvest date colocated with the large-effect QTLs for fruit loss of firmness on LG10 and LG16 (Figures 4 and 5), with the same trend towards early genotypes softening more, which confirms the close relationship between fruit ripening and softening. Using the LG10 and LG16 QTLs for selection of new varieties that will soften less during storage will thus inadvertently concurrently select for later maturing genotypes. Loss of fruit firmness and starch degradation (i.e., harvest date) are both modulated by ethylene. A model for fruit ripening based on ethylene dependency and sensitivity was developed recently.46 The model suggests that starch degradation is not dependent on ethylene; however, it is very sensitive to it, while loss of fruit firmness is strongly dependent on ethylene but not sensitive. We hypothesize that the QTLs on LG10 (PG1-RG and PG1-BB) and LG16 (Lg16-BB) could be related to variable sensitivity and dependence on ethylene. While the role of ethylene on PG1 activity has been well established,20 further research is required to identify the physiological and molecular determinism of the LG16 QTL.
Conclusions
This study highlights the need for consideration of local environmental conditions to be made when selecting new cultivars with new and improved quality traits. While the large-effect QTLs act independently from the influence of the environment, it is likely that breeding programmes will either fix these in the parents, or select them using MAS. From there, the differentiation of the new elite cultivars will be determined by small-effect QTLs that can be affected by the environment, leading to the possibility of undesirable variability of these cultivars in different environments. It also highlights the need for better molecular understanding of the processes involved in building and maintaining quality traits in apples.
References
Leeuwen CV, Roby JP, Pernet D, Bois B . Methodology of soil-based zoning for viticultural terroirs. Bull l'OIV 2010; 83: 13–29.
Lin-Wang K, Micheletti D, Palmer J et al. High temperature reduces apple fruit colour via modulation of the anthocyanin regulatory complex. Plant Cell Environ 2011; 34: 1176–1190.
Sugiura T, Ogawa H, Fukuda N, Moriguchi T . Changes in the taste and textural attributes of apples in response to climate change. Sci Rep 2013; 3: 7.
Harker FR, Gunson FA, Jaeger SR . The case for fruit quality: an interpretive review of consumer attitudes, and preferences for apples. Postharvest Biol Technol 2003; 28: 333–347.
Tacken E, Ireland H, Gunaseelan K et al. The role of ethylene and cold temperature in the regulation of the apple POLYGALACTURONASE1 gene and fruit softening. Plant Physiol 2010; 153: 294–305.
Giovannoni J . Molecular biology of fruit maturation and ripening. Annu Rev Plant Physiol Plant Mol Biol 2001; 52: 725–749.
Giovannoni JJ . Fruit ripening mutants yield insights into ripening control. Curr Opin Plant Biol 2007; 10: 283–289.
Palmer JW, Harker FR, Tustin DS, Johnston J . Fruit dry matter concentration: a new quality metric for apples. J Sci Food Agric 2010; 90: 2586–2594.
Saei A, Tustin DS, Zamani Z, Talaie A, Hall AJ . Cropping effects on the loss of apple fruit firmness during storage: the relationship between texture retention and fruit dry matter concentration. Sci Hort 2000; 130: 256–265.
Defilippi BG, Dandekar AM, Kader AA . Impact of suppression of ethylene action or biosynthesis on flavor metabolites in apple (Malus domestica Borkh) fruits. J Agric Food Chem 2004; 52: 5694–5701.
Oraguzie NC, Volza RK, Whitworth CJ et al. Influence of Md-ACS1 allelotype and harvest season within an apple germplasm collection on fruit softening during cold air storage. Postharvest Biol Technol 2007; 44: 212–219.
Goulao LF, Cosgrove DJ, Oliveira CM . Cloning, characterisation and expression analyses of cDNA clones encoding cell wall-modifying enzymes isolated from ripe apples. Postharvest Biol Technol 2008; 48: 37–51.
Ireland H, Gunaseelan K, Muddumage R et al. Ethylene regulates apple (Malus × domestica) fruit softening through a dose × time-dependent mechanism and through differential sensitivities and dependencies of cell wall-modifying genes. Plant and Cell Physiology 2014; 55 (5): 1005–1016.
Wakasa Y, Kudo H, Ishikawa R et al. Low expression of an endopolygalacturonase gene in apple fruit with long-term storage potential. Postharvest Biol Technol 2006; 39: 193–198.
Oraguzie NC, Iwanami H, Soejima J, Harada T, Hall A . Inheritance of the Md-ACS1 gene and its relationship to fruit softening in apple (Malus × domestica Borkh.). Theor Appl Genet 2004; 108: 1526–1533.
Costa F, Stella S, van de Weg WE et al. Role of the genes Md-ACO1 and Md-ACS1 in ethylene production and shelf life of apple (Malus domestica Borkh). Euphytica 2005; 141: 181–190.
Costa F, van de Weg WE, Stella S et al. Map position and functional allelic diversity of Md-Exp7, a new putative expansin gene associated with fruit softening in apple (Malus × domestica Borkh.) and pear (Pyrus communis). Tree Genet Genomes 2008; 4: 575–586.
Longhi S, Hamblin MT, Trainotti L et al. A candidate gene based approach validates Md-PG1 as the main responsible for a QTL impacting fruit texture in apple (Malus × domestica Borkh). BMC Plant Biol 2013; 13: 37.
Longhi S, Moretto M, Viola R, Velasco R, Costa F . Comprehensive QTL mapping survey dissects the complex fruit texture physiology in apple (Malus × domestica Borkh.). J Exp Botany 2012; 63: 1107–1121.
Atkinson RG, Sutherland PW, Johnston SL et al. Down-regulation of POLYGALACTURONASE1 alters firmness, tensile strength and water loss in apple (Malus × domestica) fruit. BMC Plant Biol 2012; 12: 129.
Volz RK, White AG, Brookfield PL et al. Breeding and development of the new apple Cultivar ‘Jazz’™. Acta Hort 2004; 663: 891–894.
Palmer J, Diack R, Johnston J, Boldingh H . Manipulation of fruit dry matter accumulation and fruit size in ‘Scifresh’ apple through alteration of the carbon supply, and its relationship with apoplastic sugar composition. J Hort Sci Biotechnol 2013; 88: 483–489.
Hosono S, Faruqi AF, Dean FB et al. Unbiased whole-genome amplification directly from clinical samples. Genome Res 2003; 13: 954–964.
Chagné D, Krieger C, Rassam M et al. QTL and candidate gene mapping for polyphenolic composition in apple fruit. BMC Plant Biol 2012; 12: 12.
Schaffer RJ, Friel EN, Souleyre EJ et al. A genomics approach reveals that aroma production in apple is controlled by ethylene predominantly at the final step in each biosynthetic pathway. Plant Physiol 2007; 144: 1899–1912.
Costa F, Peace CP, Stella S et al. QTL dynamics for fruit firmness and softening around an ethylene-dependent polygalacturonase gene in apple (Malus × domestica Borkh.). J Exp Botany 2010; 61: 3029–3039.
Kenis K, Keulemans J, Davey MW . Identification and stability of QTLs for fruit quality traits in apple. Tree Genet Genomes 2008; 4: 647–661.
Liebhard R, Kellerhals M, Pfammatter W, Jertmini M, Gessler C . Mapping quantitative physiological traits in apple (Malus × domestica Borkh.). Plant Mol Biol 2003; 52: 511–526.
Cevik V, Ryder CD, Popovich A et al. A FRUITFULL-like gene is associated with genetic variation for fruit flesh firmness in apple (Malus domestica Borkh.). Tree Genet Genomes 2010; 6: 271–279.
Bink MC, Jansen J, Madduri M et al. Bayesian QTL analyses using pedigreed families of an outcrossing species, with application to fruit firmness in apple. Theor Appl Genet 2014; 127: 1073–1090.
Costa F, Cappellin L, Farneti B et al. Advances in QTL mapping for ethylene production in apple (Malus × domestica Borkh.). Postharvest Biol Technol 2014; 87: 126–132.
Costa F, Cappellin L, Farneti B et al. Texture dynamics during postharvest cold storage ripening in apple (Malus × domestica Borkh.). Postharvest Biol Technol 2012; 69: 54–63.
Costa F, Cappellina L, Longhia S et al. Assessment of apple (Malus × domestica Borkh.) fruit texture by a combined acoustic–mechanical profiling strategy. Postharvest Biol Technol 2011; 61: 21–28.
King GJ, Lynn JR, Dover CJ, Evans KM, Seymour GB . Resolution of quantitative trait loci for mechanical measures accounting for genetic variation in fruit texture of apple (Malus pumila Mill.). Theor Appl Genet 2001; 102: 1227–1235.
King GJ, Lynn JR, Dover CJ et al. Quantitative genetic analysis and comparison of physical and sensory descriptors relating to fruit flesh firmness in apple (Malus pumila Mill.). Theor Appl Genet 2000; 100: 1074–1084.
Nybom H, Afzadi MA, Sehic J, Hertog M . DNA marker-assisted evaluation of fruit firmness at harvest and post-harvest fruit softening in a diverse apple germplasm. Tree Genet Genomes 2013; 9: 279–290.
Beavis WD, Smith OS, Grant D, Fincher R . Identification of quantitative trait loci using a small sample of topcrossed and F4 progeny from maize. Crop Sci 1994; 34: 882–896.
Nardozza S, Boldingh HL, Osorio S et al. Metabolic analysis of kiwifruit (Actinidia deliciosa) berries from extreme genotypes reveals hallmarks for fruit starch metabolism. J Exp Botany 2013; 64: 5049–5063.
Quilot B, Kervella J, Genard M, Lescourret F . Analysing the genetic control of peach fruit quality through an ecophysiological model combined with a QTL approach. J Exp Botany 2005; 56: 3083–3092.
Prudent M, Causse M, Génard M, Tripodi P, Grandillo S, Bertin N . Genetic and physiological analysis of tomato fruit weight and composition: influence of carbon availability on QTL detection. J Exp Botany 2009; 60: 923–937.
Velasco R, Zharkikh A, Affourtit J et al. The genome of the domesticated apple (Malus × domestica Borkh.). Nat Genet 2010; 42: 833.
Kumar S, Garrick DJ, Bink MC, Whitworth C, Chagné D, Volz RK . Novel genomic approaches unravel genetic architecture of complex traits in apple. BNC Genomics 2013; 14: 393.
Zhu YM, Barritt BH . Md-ACS1 and Md-ACO1 genotyping of apple (Malus × domestica Borkh.) breeding parents and suitability for marker-assisted selection. Tree Genet Genomes 2008; 4: 555–562.
Atkinson RG, Bolitho KM, Wright MA, Iturriagagoitia-Bueno T, Reid SJ, Ross GS . Apple ACC-oxidase and polygalacturonase: ripening-specific gene expression and promoter analysis in transgenic tomato. Plant Mol Biol 1998; 38: 449–460.
Johnston JW, Hewett EW, Hertog M, Harker FR . Harvest date and fruit size affect postharvest softening of apple fruit. J Hort Sci Biotechnol 2002; 77: 355–360.
Johnston JW, Gunaseelan K, Pidakala P, Wang M, Schaffer RJ . Co-ordination of early and late ripening events in apples is regulated through differential sensitivities to ethylene. J Exp Botany 2009; 60: 2689–2699.
Acknowledgements
This research was funded by a New Zealand Ministry of Business, Innovation and Employment (MBIE) grant: ‘Pipfruit: a Juicy Future’ (C06X0705). We thank Dr Massimo Pindo, Dr Michela Troggio and Dr Riccardo Velasco from Fondazione Edmunch Mach, Italy, and Dr Baptiste Guitton for the SNP genotyping, and Andrew McLachlan and Nihal de Silva for help with the statistical analysis.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivs 3.0 Unported License. The images or other third party material in this article are included in the article's Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/3.0/
About this article
Cite this article
Chagné, D., Dayatilake, D., Diack, R. et al. Genetic and environmental control of fruit maturation, dry matter and firmness in apple (Malus × domestica Borkh.). Hortic Res 1, 14046 (2014). https://doi.org/10.1038/hortres.2014.46
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1038/hortres.2014.46