Abstract
A common disorder of human dentition is the existence of supernumerary teeth. Impacted supernumerary teeth occur most frequently in the maxillary incisor area and are termed mesiodens. We conducted whole-exome sequencing of non-syndromic Japanese individuals possessing supernumerary teeth to identify genes and/or loci involved in the pathogenesis of the condition.
Similar content being viewed by others
Teeth are an important vertebrate organ, especially in humans because of their use in feeding, mastication, and speech. Tooth development and formation involve various complex processes, including epithelial-mesenchymal transitions, that are precisely controlled by molecular signaling pathways at each developmental stage.1 A common disorder found in human dentition is the existence of supernumerary teeth. There are 20 teeth in the primary dentition and 28 teeth, excluding third molars, in the permanent dentition. Supernumerary teeth are defined as extra teeth—more than the normal human complement.2 The frequency of this condition in permanent dentition ranges from approximately 0.1 to 3.6%, with varied ranges among populations,3 and is more prevalent among males than females, with a ratio of approximately 2:1.4 Supernumerary teeth are classified by their various morphologies or location. The most common supernumerary teeth are located in the maxillary incisor area and are called mesiodens.5 Mesiodens are typically conical supernumerary teeth that are small and peg-shaped. The autosomal-dominant transmission pattern of non-syndromic multiple supernumerary teeth has been reported.6
Cleidocranial dysplasia (CCD) is a human syndrome that presents with supernumerary teeth. It is an autosomal-dominant disorder characterized by short stature, hypoplasia or aplasia of clavicles, delayed closure of the cranial fontanelles and sutures, maxillary hypoplasia, Wormian bones, frontal bossing, wide pubic symphysis, and dental abnormalities including supernumerary teeth.7 Mutations in runt related transcription factor 2 (RUNX2), an essential transcription factor for osteoblast differentiation and skeletal development8, have been identified as a main cause for CCD.9 In addition to CCD, other human syndromes are also associated with supernumerary teeth.10
Various mouse models have been used to explore the etiology of supernumerary teeth and to identify candidate genes involved in the pathogenesis of this condition. Supernumerary teeth in the diastema region in front of the first molars are exhibited by Sprouty2 and Sprouty4 null mice (fibroblast growth factor (Fgf) antagonists), Tabby mice, which have a mutation in ectodysplasin (Eda), transgenic mice overexpressing Eda or ectodysplasin receptor (Edar), paired box 6 (Pax6) mutant mice, and growth arrest specific 1 (Gas1) null mutant mice. Furthermore, supernumerary teeth in both incisor and molar regions are exhibited by specificity protein 6 (Sp6) deficient mice and sclerostin domain containing 1 (Sostdc1) (also known as ectodin, Wise and USAG-1) null mutant mice.10 Mice possessing conditional knockout of the adenomatous polyposis coli (Apc) gene under the control of the human keratin 14 (K14) promoter also exhibited supernumerary teeth. This study indicated that the occurrence of supernumerary teeth by loss of Apc was through activation of Wnt/β-catenin signaling.11 The normal number of teeth in mice is, however, less than that of humans, and their teeth are not replaced because they have only primary dentition. Therefore, mouse models may not be the most appropriate ones for determining the etiology of supernumerary teeth in humans.
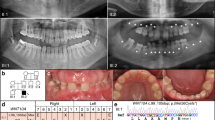
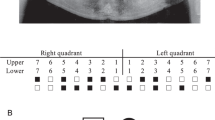
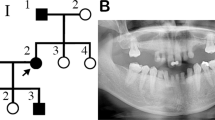
In the present study, we selected four Japanese families, Families A, B, C, and D. In three families (Families A, B and D), both first and second generations have supernumerary teeth. We do not know whether the first generation in Family C is affected. However, the second generation in Family C has impacted supernumerary teeth in the maxillary incisor area. Most affected individuals have one supernumerary tooth; however, II-2 in Family A has two (Figure 1a, b). The occurrence of supernumerary teeth in most of these families indicates an autosomal-dominant pattern of inheritance. All subjects were diagnosed by examining a panoramic radiograph or upper anterior occlusal radiographs and interviewing the patient about their medical and dental history. All subjects were free of any syndrome or congenital anomaly such as cleft lip or palate. All subjects provided written informed consent and the study was approved by the Ethics Committee of Showa and Kanazawa University.
(a) Upper anterior occlusal radiograph of II-2 in Family A with two impacted supernumerary teeth in the maxillary incisor area. (b) Family pedigrees of Japanese individuals with supernumerary teeth. Squares and circles denote males and females, respectively. Filled symbols indicate affected individuals. The phenotype of the first generation in Family C is unknown.
Saliva samples were collected from 16 individuals (10 affected and 6 unaffected). DNA was extracted from the samples using Oragene DNA (DNA Genotek, Ottawa, Canada) and subjected to whole-exome sequencing. DNA samples (3 μg) were subjected to exome capture using the Sure-Select Human All Exon Kit (Agilent Technologies, Santa Clara, CA, USA) according to the manufacturer’s instructions. The captured DNA underwent high-throughput sequencing using the HiSeq2500 system (Illumina, San Diego, CA, USA). All prepared flow cells were run on the Illumina HiSeq2500 using paired-end 100-bp reads. Reads were mapped to the reference genome (UCSC hg19) using the Burrows-Wheeler Aligner (BWA) v.0.7.9.12 BWA-generated SAM files were sorted and indexed using SAMtools v.0.1.18.13 The files obtained in BAM format were analyzed using GATK v.2.7 following their best practice guidelines.14 To perform gene annotation, we used the RefSeq gene database (build hg19),15 while variant annotation was based on the dbSNP (dbSNP 135), the 1000 Genomes Project database,16 and data of 1,208 Japanese individuals from the Human Genetic Variation database (HGVD) (http://www.genome.med.kyoto-u.ac.jp/SnpDB/index.html). In all family samples, variants detected in exome sequencing data were further analyzed by performing three filtering steps based on different criteria. Rare variants with frequencies <3% in the Japanese population were selected from HGVD to obtain rare variants as potential candidates.
Whole-exome sequencing identified 149 single-nucleotide variants (SNVs) and seven small insertions and deletions (indels) that co-segregated in Family A in the fully penetrant autosomal-dominant mode of inheritance. Moreover, 157 SNVs and seven indels co-segregated in Family B, 407 SNVs and 28 indels co-segregated in Family C, and 16 SNVs and four indels co-segregated in Family D. However, we did not identify any common gene variants in all four families. Three families (Families A, C, and D) all had variants in SCO-spondin (SSPO) (Table 1). It is known that SSPO plays a role in central nervous system development.17,18 We detected 23 common gene variants in two families. Families A and B both had variants in EF-hand calcium binding domain 5 (EFCAB5), testis expressed 15 (TEX15), and tumor suppressor candidate 1 (TUSC1). Families A and C both had variants in absent in melanoma 1-like (AIM1L), cadherin 26 (CDH26), exocyst complex component 3-like 4 (EXOC3L4), family with sequence similarity 186 member A (FAM186A), FXYD domain-containing ion transport regulator 4 (FXYD4), polycystin 1 like 2 (PKD1L2), LOC100652824, and transketolase-like 1 (TKTL1). Families B and C both had variants in agrin (AGRN), ataxin 1 (ATXN1), chromosome 21 open reading frame 58 (C21orf58), complement factor B (CFB), Fanconi anemia complementation group E (FANCE), formin-like 1 (FMNL1), hemicentin 1 (HMCN1), immunoglobulin superfamily member 9B (IGSF9B), KIAA1614, MGA, MAX dimerization protein (MGA), phospholipase C eta 2 (PLCH2), and ring finger protein 207 (RNF207) (Table 2). Up to date, none of these genes are known to have a function in tooth development.
This is the first study to demonstrate the genetic landscape in non-syndromic Japanese patients with impacted supernumerary teeth in the maxillary incisor area. Whole-exome sequencing is an effective strategy to elucidate the etiology of dental diseases, and our findings provide a basis for further exploration of the pathological mechanisms of supernumerary teeth.
References
References
Marianna B . Molecular genetics of tooth development. Curr Opin Genet Dev 2009; 19: 504–510.
Schulze C Developmental abnormalities of the teeth and jawsIn: Gorlin RJ, Goldman HM (eds) Thomas’ Oral Pathology 6th edn, Vol. 1. Mosby: St Louis, USA, 1970, pp 96-183.
Yusof WZ . Non-syndromic multiple supernumerary teeth: literature review. J Can Dent Assoc 1990; 56: 147–149.
Rajab LD, Hamdan MA . Supernumerary teeth: review of the literature and a survey of 152 cases. Int J Paediatr Dent 2002; 12: 244–254.
Hyun HK, Lee SJ, Lee SH, Hahn SH, Kim JW . Clinical characteristics and complications associated with mesiodentes. J Oral Maxillofac Surg 2009; 67: 2639–2643.
Batra P, Duggal R, Parkash H . Non-syndromic multiple supernumerary teeth transmitted as an autosomal dominant trait. J Oral Pathol Med 2005; 34: 621–625.
Mundlos S . Cleidocranial dysplasia: clinical and molecular genetics. J Med Genet 1999; 36: 177–182.
Komori T, Yagi H, Nomura S, Yamaguchi A, Sasaki K, Deguchi K et al. Targeted disruption of Cbfa1 results in a complete lack of bone formation owing to maturational arrest of osteoblasts. Cell 1997; 89: 755–764.
Mundlos S, Otto F, Mundlos C, Mulliken JB, Aylsworth AS, Albright S et al. Mutations involving the transcription factor CBFA1 cause cleidocranial dysplasia. Cell 1997; 89: 773–779.
Wang XP, Fan J . Molecular genetics of supernumerary tooth formation. Genesis 2011; 49: 261–277.
Wang XP, O’Connell DJ, Lund JJ, Saadi I, Kuraguchi M, Turbe-Doan A et al. Apc inhibition of Wnt signaling regulates supernumerary tooth formation during embryogenesis and throughout adulthood. Development 2009; 136: 1939–1949.
Li H, Durbin R . Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009; 25: 1754–1760.
Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009; 25: 2078–2079.
DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet 2011; 43: 491–498.
Pruitt KD, Brown GR, Hiatt SM, Thibaud-Nissen F, Astashyn A, Ermolaeva O et al. RefSeq: an update on mammalian reference sequences. Nucleic Acids Res 2014; 42: D756–D763.
Abecasis GR, Auton A, Brooks LD, DePristo MA, Durbin RM, Handsaker RE et al. An integrated map of genetic variation from 1,092 human genomes. Nature 2012; 491: 56–65.
Gobron S, Creveaux I, Meiniel R, Didier R, Herbet A, Bamdad M et al. Subcommissural organ/Reissner’s fiber complex: characterization of SCO-spondin, a glycoprotein with potent activity on neurite outgrowth. Glia 2000; 32: 177–191.
Meiniel A . SCO-spondin, a glycoprotein of the subcommissural organ/Reissner’s fiber complex: evidence of a potent activity on neuronal development in primary cell cultures. Microsc Res Tech 2001; 52: 484–495.
Data Citations
Yamaguchi, Tetsutaro HGV Database (2016) http://dx.doi.org/10.6084/m9.figshare.hgv.938
Yamaguchi, Tetsutaro HGV Database (2016) http://dx.doi.org/10.6084/m9.figshare.hgv.941
Yamaguchi, Tetsutaro HGV Database (2016) http://dx.doi.org/10.6084/m9.figshare.hgv.944
Yamaguchi, Tetsutaro HGV Database (2016) http://dx.doi.org/10.6084/m9.figshare.hgv.947
Acknowledgements
This work was supported by JSPS KAKENHI (16K15843; Grant-in-Aid for challenging Exploratory Research to TY) and by the Ministry of Education, Culture, Sports, Science and Technology (MEXT)—Supported Program for the Strategic Research Foundation at Private Universities, 2012–2017.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/4.0/
About this article
Cite this article
Takahashi, M., Hosomichi, K., Yamaguchi, T. et al. Whole-exome sequencing analysis of supernumerary teeth occurrence in Japanese individuals. Hum Genome Var 4, 16046 (2017). https://doi.org/10.1038/hgv.2016.46
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1038/hgv.2016.46
This article is cited by
-
Association of TUSC1 and DPF3 gene polymorphisms with male infertility
Journal of Assisted Reproduction and Genetics (2018)