Abstract
It has been suggested that adaptive evolution on ecological timescales shapes communities. However, adaptation among environments relies on isolation or large selection coefficients that exceed migration effects. This reliance is tempered if adaptation is polygenic—does not depend on one allele completely replacing another but instead requires small allele frequency changes at many loci. Thus, whether individuals can evolve adaptation to fine-scale habitat variation (for example, microhabitats) is not resolved. Here we analyze the genetic divergence of the teleost fish, Fundulus heteroclitus, among microhabitats that are <200 m apart in three separate saltmarshes using 4741 single-nucleotide polymorphisms (SNPs). Among these SNPs, 1.3–2.3% have large and highly significant differences among microhabitats (mean FST=0.15; false discovery rate ⩽1%). The divergence among microhabitats for these outlier SNPs is larger than that among populations, exceeds neutral expectation and indicates surprising population structure among microhabitats. Thus, we suggest that polygenic selection is surprisingly effective in altering allele frequencies among many different SNPs that share similar biological functions in response to environmental and ecological differences over very small geographic distances. We acknowledge the evolutionary difficulty of large genetic divergence among well-connected habitats. Therefore, these studies are only the first step to discern whether natural selection is responsible and capable of effecting genetic divergence on such a fine scale.
Similar content being viewed by others
Introduction
Individuals should be adapted to their local environment to maximize their fitness (Williams, 1966). However, adaptation among environments relies on isolation or large selection coefficients that exceed migration effects (Slatkin, 1987; Nielsen et al., 2009). This reliance is tempered if adaptation does not depend on one allele completely replacing another (Otto and Whitlock, 1997) but instead requires small allele frequency changes at many loci (Turelli and Barton, 2004; Przeworski et al., 2005; Pritchard et al., 2010). Adaptive changes involving many loci likely require significant standing genetic variation (Bergland et al., 2014). Yet, this creates a paradox: more genetic variation is being detected in natural populations than would be expected given our theoretical understanding of population genetics (Mackay et al., 2012; Corander et al., 2013; Hess et al., 2013; Messer and Petrov, 2013; Bergland et al., 2014; Huang et al., 2014; Charlesworth, 2015). Several recent studies suggest that this copious standing genetic variation could be a significant resource for adaptation (Przeworski et al., 2005; Messer and Petrov, 2013). The paradox of ‘too much’ polymorphism was initially noted in the landmark paper of Lewontin and Hubby (1966) on the frequency of protein polymorphisms. With ∼30% of proteins having polymorphisms, the authors concluded that a large amount of standing genetic variation existed, but they lacked a biological mechanism that could maintain the observed level of variation.
One explanation is that genetic polymorphisms exist as transient changes due to neutral drift (Kimura, 1968). This theory, in many ways, solved the problem of genetic load: the cost of too many selective deaths if many genetic polymorphisms were being affected by adaptive evolution (Haldane, 1957; Crow, 1958). The neutral theory undoubtedly correctly explains many, if not most, nucleotide variation patterns. Yet, this elegant and parsimonious theory often fails to explain observed nucleotide variation patterns (Ohta, 1992; Kreitman, 1996). A recent example is the apparent adaptive variation of hundreds of single-nucleotide polymorphisms (SNPs) in response to seasonal environmental oscillations within a Drosophila melanogaster population (Bergland et al., 2014). These data and other genome-wide studies have revealed genetic variation within and among populations of Drosophila, humans and other organisms that is greater than is predicted by the neutral theory (Mackay et al., 2012; Corander et al., 2013; Hess et al., 2013; Messer and Petrov, 2013; Bergland et al., 2014; Huang et al., 2014; Charlesworth, 2015; Henn et al., 2015; Yeaman, 2015).
Until recently, few opportunities existed to determine the number or frequency of adaptive versus neutral variation patterns. The biological importance of genome-wide variation (versus focusing on one or a few genes, for example, microsatellites) is supported by the D. melanogaster Genetic Reference Panel (DGRP) where the significant genotypic variation within a population is associated with fitness-related traits such as oxidative stress response, chill coma recovery and starvation resistance (Mackay et al., 2012; Huang et al., 2014; Charlesworth, 2015). Adaptive divergence was also demonstrated in the Baltic Sea herring, Clupea harengus (Corander et al., 2013), and Pacific lamprey, Entosphenus tridentatus (Hess et al., 2013), based on significant population structure with a few hundred SNPs that were not evident based on thousands of neutral markers. These data in Drosophila, herring and other species indicate that hundreds, and sometimes thousands, of SNPs across the genome are affected by natural selection (Bergland et al., 2014; Yeaman, 2015). Yet, these observations raise the old problem concerning the number of SNPs affected by selection—excessive genetic load (Haldane, 1957; Crow, 1958; Henn et al., 2015). The theoretical understanding of how large amounts of standing genetic variation can contribute to adaptation while not conferring a lethal level of genetic load has not been resolved. To provide insight into this problem requires large populations that are likely to be affected by differential selective pressure related to environmental variation.
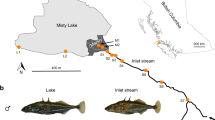
The teleost fish Fundulus heteroclitus has been a model of evolved changes within and among populations because of its well-studied natural history, ecology, biochemistry and molecular genetics (Newman, 1907; Kneib, 1986; Powers et al., 1991; Hunter et al., 2007; Duvernell et al., 2008). F. heteroclitus have large populations exceeding 10 000 individuals (Duvernell et al., 2008) within a single Spartina saltmarsh. Within these saltmarshes, daily tides flush individuals in and out of the estuary (Able et al., 2012); in addition, individuals breed and lay eggs in a the upper intertidal zone (Newman, 1907), suggesting well-mixed populations (Newman, 1907; Kneib, 1986; Able et al., 2012). Yet, within a single saltmarsh are three microhabitats: (1) tidal basins (B), (2) creeks (C) and (3) ponds (P), with meaningful environmental differences in daily maximum temperatures, oxygen concentrations and productivity (Hunter et al., 2007). Mark–recapture studies suggest individuals are often associated with a single microhabitat and small home range (<200 m; Skinner et al., 2012). To explore the effect of microhabitat (Basin, Creek and Pond) on genetic structure within and among estuaries, we examined three separate F. heteroclitus populations along the North American New Jersey coast: Mantoloking (Mk), Rutgers University Marine Field Station (RM) and Stone Harbor (SH; Figure 1). We used genotyping by sequencing (GBS, (Elshire et al., 2011)) to generate 4741 SNPs (with 5% minor allele frequency (MAF) and not in linkage disequilibrium) and analyzed the genetic variation within and among microhabitats.
Sampling locations along the New Jersey coast (Mantoloking (Mk), Rutgers University Marine Field Station (RM) and Stone Harbor (SH)). An expanded image of Rutgers University Marine Field Station site with basin (B), creek (C) and pond (P) microhabitats identified. The distance between microhabitats was never >200 m and usually <50 m.
We present analyses that demonstrate that 1.3–2.3% of 4741 SNPs in three separate saltmarshes have large and highly significant differences among microhabitats that are <200 m apart. These divergent outlier SNPs indicate surprising population structure among microhabitats most likely due to fine-scale adaptive divergence.
Materials and methods
Study species and study sites
F. heteroclitus (n=215) were collected from three estuarine habitats along the New Jersey coast, USA (Figure 1) between 1 and 15 July 2013. Collection sites included Mantoloking, NJ (Mk; 40°02’59.07' N, 74°04'08.64' W; n=61), Rutgers University Marine Field Station, NJ (RM; 39°30’31.34' N, 74°19’27.12' W; n=92) and Stone Harbor, NJ (SH; 39°03’46.46' N, 74°46’42.90' W; n=62). Individuals were captured in three distinct microhabitat types: tidal basins, intertidal creeks and permanent ponds. Within each estuary, the distance from one microhabitat to another was typically <50 m.
Sample collection
Fish were captured in baited minnow traps during incoming tides. Fish were kept in a shaded bucket with aerated sea water until processing (<1 h). Tissue from each individual was nonlethally collected via caudal fin clip; then the fish was released at the capture location. Fin clips were immediately immersed in Chaos buffer (4.5 M guanidinium thiocyanate, 2% N-lauroylsarcosine, 50 mM EDTA, 25 mM Tris-HCL, pH 7.5, 0.2% antifoam and 0.1 M β-mercaptoethanol) and then put on ice within 1 h of collection. Tissue samples were then stored at −20 °C before processing.
Genomic DNA isolation
Genomic DNAs from fin clips were isolated using silica columns (Ivanova et al., 2006). Genomic DNA quality was assessed via gel electrophoresis, and concentrations were quantified using Biotium AccuBlue Broad Range dsDNA Quantitative Solution kit (Fremont, CA, USA) according to the manufacturer’s instructions. Then, 100 ng of genomic DNA from each sample was dried down in a 96-well plate. Samples were hydrated overnight with 5 μl of water before further processing.
Genotyping by sequencing
The GBS protocol described in Elshire et al. (2011) was used to generate an Illumina sequencing library using the restriction enzyme AseI, adaptors (0.4 pmol per sample) and 50 ng of genomic DNA. This library was sequenced on Illumina HiSeq 2500 (San Diego, CA, USA) with a 75 bp single-end read (Elim Biopharmaceuticals, Inc., Hayward, CA, USA).
Sequence alignment and SNP filtering
The reference genome-based GBS pipeline TASSEL ver. 4.0 (Bradbury et al., 2007) was used to identify SNPs using the F. heteroclitus genome assembly (Reid et al., 2015). Bowtie2 was used to align reads to the F. heteroclitus genome. The resulting alignment and respective SNP identifications yielded 338 325 SNPs. SNPs were filtered so that each individual had calls for 30% of all SNPs and each SNP was called in 80% of individuals, leaving 15 259 SNPs in 202 individuals. That is, 13 individuals were excluded because these individuals had too few SNPs shared with other individuals.
Linkage disequilibrium and distance among SNPs
To determine the linkage groups and distances among the 15 259 SNPs, we used a 50-SNP moving window to calculate r2 and relative disequilibrium, D’ (Lewontin, 1964), using TASSEL (Bradbury et al., 2007). Distances among all SNPs (with or without significant linkage disequilibrium (LD)) within a scaffold were calculated, resulting in the maximum distance among 50 SNPs equal to >106 bp. Significant LDs were defined as r2<0.05 and D’ P-values of <0.01, without multiple correction to avoid type II errors (mistakenly accepting the null hypothesis of no LD).
Filtering to identify independent orthologous SNPs
We filtered the SNP to only those with a 5% minimum allele frequency and not in significant LD with other SNPs (r2<0.05 and P<0.01). We excluded SNPs that had observed heterozygosity (Ho) significantly larger than expected heterozygosity (He, that is, Ho>>He, P<0.01) using Arlequin ver. 3.5.1.2 (Excoffier and Lischer, 2010). Using these criteria left 4741 SNPs in 202 individuals. A 5% minimum allele frequency insures that at least 10 individuals had the minor allele. Exclusion of SNPs with large significant Ho excludes nucleotide variation created by alignment between paralogs (versus true allelic variation among orthologs; Nunez et al., 2015; Crawford and Oleksiak, 2016). Selection of unlinked SNPs reduced bias due to background selection or selective sweeps. Further statistical comparisons considered only these 4741 SNPs.
Statistic analyses and experimental design
There are two types of analyses: (1) analyses considering all 4741 SNPs together to estimate overall population differentiation, and (2) analyses of each SNP separately to define neutral and informative SNPs. We analyzed all SNPs together to determine the overall population structure. Analyses using all 4741 SNPs together used pairwise comparisons between microhabitats within and among populations and avoided the potential effects of a minority of SNPs with nonneutral divergence effecting measures of population structure.
We also examined each SNP separately to define SNPs with large genetic differences among microhabitat where none should exist if the population is panmictic. These analyses define which SNPs are associated with population structure among microhabitats. Analyses of individual SNPs did not use pairwise comparisons; instead individual SNP analyses define genetic distance among all three microhabitats (versus three separate pair comparisons). This approach was taken to avoid multiple comparisons. To examine the SNP-specific differences among microhabitats, we employed three tests: SNP-specific FST and two common outlier tests (LOSITAN and Arlequin, see below). We use these three tests because each test has different statistical and evolutionary strengths and weaknesses.
Overall population FST
Pairwise differences within and among populations were calculated using ‘ape’ (R-package (Paradis et al., 2004), ver. 4.0). Hierarchical, locus-by-locus, analyses of molecular variance (AMOVA; Excoffier et al., 1992) were performed with Arlequin using 50 000 permutations (P<0.01); comparisons were made among populations and nested microhabitats (basin, creek and pond) within their respective estuary population (Mk, RM and SH). The locus-by-locus model was used to weight the SNPs appropriately to account for variation in the degrees of freedom per individual (Mace et al., 2006; Excoffier and Lischer, 2010; Lind and Grahn, 2011).
Significant SNP-specific FST permutation test
We calculated SNP-specific FST values, and for each SNP we compared this original FST value with a distribution of values for that SNP when individuals were randomly shuffled among microhabitats. This compares FST values for each SNP only to randomized values of the same SNP and ignores the global distribution of the FST values of other SNPs. The FST values for each SNP among the three microhabitats within each population were calculated using the Weir–Cockerham FST estimator (Weir and Cockerham, 1984). In order to estimate a random FST value distribution for each SNP separately within each estuary population, individuals were randomly assigned to basin, creek and pond, and FST values were calculated 1000 × using the Weir–Cockerham FST estimator in ‘vcftools’ (Danecek et al., 2011). These random values were used in a cumulative distribution function in R to estimate the probability of achieving the original FST values. For each original SNP, the FST values were deemed significantly different (P<0.01) if they occurred in 1% of the randomly permutated values for that SNP.
SNP-specific neutrality tests
LOSITAN ver. 1.0 (Beaumont and Nichols, 1996; Antao et al., 2008) was used to estimate genetic diversity among the individuals and populations collected by calculating the percentage of polymorphic SNPs observed (Ho) and expected (He), as well as the fixation index (FST). LOSITAN was then used to define SNPs with outlier FST values (larger than expected FST values relative to a permutation of SNPs with similar He, P<0.01; (Beaumont and Balding, 2004)). The random expected FST values were calculated by generating 50 000 simulations that were used to estimated P-values for each SNP relative to all other SNPs with similar He (see Supplementary Figure 2). That is, LOSITAN performs 50 000 permutations of the data to provide the probability of achieving the original FST value relative to other SNPs with similar He. All comparisons with identical parameters to those used in the LOSITAN analysis were repeated using Arlequin. To determine significance in Arlequin, a coalescent simulation was used to estimate a null distribution and confidence intervals around the observed values and then tested to determine whether observed locus-specific FST values can be considered as outliers conditioned on the global observed FST value (Excoffier and Lischer, 2010). Like LOSITAN, Arlequin estimates the likelihood of a SNP-specific FST value relative to the values of other SNPs. In both analyses, the significance of the original FST value is relative to the FST value among all SNPs.
Outlier SNPs
‘Outlier SNPs’ used in subsequent analyses and presented in figures are SNPs that were significant in all three analyses (LOSITAN, Arlequin and the permutation test; P<0.01) and where the joint probability of the three analyses had a false discovery rate (FDR) of ⩽1% (Figure 2). FDR was computed via the Benjamini–Hochberg (Benjamini and Hochberg, 1995) procedure using ‘p.adjust’ in R (ver. 3.2.3).
Evolutionary analyses among microhabitats for three populations, where each population has three analyses: (1) SNPs with significantly different FST values, (2) LOSITAN-identified significant outlier SNPs and (3) Arlequin-identified significant outlier SNPs. Significant SNPs detected in all three analyses with joint FDR <1% were considered in subsequent analyses.
Neutral SNP FST distribution
To plot and compare the neutral FST value distribution relative to outlier SNP FST values, we selected neutral SNPs as those with LOSITAN P-values of >0.1 for any single population comparison (1636 SNPs). In addition, a random, neutral, FST value distribution was approximated by randomizing the microhabitat assignments within each estuary population 100 × and analyzing these permutations in LOSITAN with identical parameters as above. For each estuary population, this generated 100 estimates of neutral FST value distributions. Notice that the permutation used LOSITAN to determine significance relative to SNPs with similar He.
Discriminant analysis of principal components (DAPC)
DAPC (Jombart et al., 2010) in the R-package ‘adegenet’ (ver. 2.0.1; Jombart, 2008; Jombart and Ahmed, 2011) was used to visualize demographic relationships among microhabitats and populations. The number of microhabitats being compared defined group number prescribed to DAPC. The number of principle components prescribed to DAPC was one-third the number of individuals being compared.
Gene ontology
To annotate SNPs and provide variant type (coding, 3′ untranslated region, intronic, intergenic and so on) ‘snpEff’ (ver. 4.3i) was used with the F. heteroclitus genome (Reid et al., 2015). Because the Fundulus genome has been update recently (National Center for Biotechnology Information (NCBI): GCA_000826765.1 Fundulus_heteroclitus-3.0.2), we realigned SNP tag sequences to confirm scaffold and position using BWA (ver. 7.15; Li and Durbin, 2009). Further confirmation was achieved by BLAST alignment of tag sequences against the F. heteroclitus reference genome. Gene symbol, protein and mRNA accession were used to identify human homologs with NCBI HomoloGene. To enhance the discovery of finding human homologs, BLAST alignments were used. Human gene symbols were used with PANTHER (Mi et al., 2016) to assign Gene Ontology (GO) terms and identify statistically significant (P<0.05) overrepresentation. Only GO-SLIM terms were used.
Ethical statement
Fieldwork was completed within publically available lands, and no permission was required for access. F. heteroclitus does not have endangered or protected status, and small marine minnows do not require collecting permits for noncommercial purposes. All fish were captured in minnow traps with little stress and returned in <1 h. The procedures approved by the institutional animal care and use committee were used for sampling.
Results
SNP allele frequencies were measured in three New Jersey F. heteroclitus populations (Figure 1). Each population was stratified into the three microhabitats (B, basins; C, intertidal creeks; and P, permanent ponds; Table 1) for a total of nine unique sampling locations.
Sequences
Sequencing resulted in 144 389 606 independent, barcoded, sequences (Tags) from 215 individuals. This yielded an average coverage of 11.1 reads per individual per Tag, and a range from 7 to 464 reads per individual per Tag for the SNPs used for analysis. The following sequence alignment to the F. heteroclitus genome (Reid et al., 2015) yielded 146 431 unique 64 bp Tags; of these, 105 443 contained polymorphic sites (72.01%) and 27 264 were invariant. From these Tags, a total of 338 325 SNPs was identified with an average of 2.29 SNPs per 64 bp Tag, 3.37 SNPs per 100 bp and 1276.2 reads per Tag.
LD and distance among SNPs
We calculated the distance and LD among 15 259 SNPs using a 50-SNP window (Supplementary Figure 1a). Of the 66 853 distances among SNPs within a scaffold, 49 082 (73%) are >1000 bp apart, and 94% (16 705) of the remaining distances among SNPs are <100 bp apart. This bimodal distribution reflects GBS methods where SNPs are identified among 100 bp tag sequences that share a restriction site (two tags with a shared site=200 bp), and the distance among restriction sites is typically >1000 bp. Thus, SNPs are either within 100–200 bp or >1000 bp (Supplementary Figure 1b). Of the 8180 SNPs with significant LD (r2 <0.05 and D’ with P<0.01), 7623 (93%) are <100 bp apart (Supplementary Figure 1c). Only 401 (4.9%) SNPs in significant LD with another SNP are >1000 bp distant (Supplementary Figures 1b and c). The short LD distances are consistent with other published results of LD in F. heteroclitus (Baris et al., 2016). SNPs with significant LD (r2 <0.05 and P<0.01) were not included in any subsequent analysis.
To avoid making determination with nonindependent SNPs (that is, those in LD), SNPs that occurred in too few individuals and SNPs with a bias in one microhabitat or among paralogs, SNPs were conservatively filtered. We required that each individual had calls for 30% of all SNPs, and each SNP was called in 80% of individuals, SNPs had a 5% minimum allele frequency, SNPs did not have large observed heterozygosity (Ho) that significantly deviated from Hardy–Weinberg equilibrium expected heterozygosity (He; Ho>>He, P<0.01) and LD was not significant (r2 <0.05 and P>0.01). This produced 4741 SNPs in 202 individuals. These 4741 SNPs were used in all subsequent analyses. Thirteen individuals (primarily from RM; Table 1) were excluded because they had few called SNPs; this most likely reflects the DNA quality from these individuals. Reduction in sample size in an arbitrary set of individuals would only reduce our power to distinguish among groups and not bias the data. A potential greater concern is the reduction from >300 000 SNPs to 4741 SNPs. Most of this reduction (>300 000 SNPs to 15 259) is the requirement that SNPs are shared among 80% of all individuals. This does not eliminate rare SNPs or SNPs within any one group; it only eliminates the loci that occur in too few individuals. The lack of shared SNPs among individuals is likely due to restriction enzyme site polymorphisms that only occur in a few individuals that represent a different type of genetic variation (restriction fragment length polymorphism) not examined here. However, the minimum 5% allele frequency (5% MAF) does remove all SNPs with low MAFs. Not examining these SNPs means we are missing subtle differences at rare alleles, but it does not affect or bias the locus-by-locus (SNP-specific) analyses. Requiring each SNP to have Ho that is not significantly larger than He reduces the likelihood of comparing paralogs (Nunez et al., 2015; Crawford and Oleksiak, 2016). Requiring SNPs that are not in LD prevents the inflation of the number of significant genes. These filtering steps should not create a bias in defining differences among microhabitats or populations because they represent SNPs with good coverage that are most likely orthologs. Thus, these filters may reduce the power of analyses, but they should not create an ascertainment bias.
Analysis of molecular variance
The genetic variation among all 4741 SNPs was examined with a nested AMOVA (Excoffier et al., 1992) that compares the genetic variation across all SNPs among microhabitats relative to the variation among individuals and the variation among populations relative to the variation among microhabitats (Table 2). Across all SNPs, most variation, >98%, occurs among individuals within a microhabitat. One percent of the variation occurs between microhabitats, and a small (~0.1%), but significant, portion of the variation occurs between estuary populations (Mk, RM and SH; Table 2). These data suggest that there are small but significant genetic differences between the three NJ estuary populations and, surprisingly, an order of magnitude more variance between microhabitats within a single estuary population than among populations.
Fst values using all 4741 SNPs
To determine the genome-wide genetic divergence we calculated FST values using all 4741 SNPs (Table 3). FST values contrast the genetic variance among groups relative to the total variance (total=within plus between), and the significance is determined by random permutation of individuals into groups with equal sample sizes. The FST values across all populations (Mk vs RM vs SH) are small but significant (FST=0.00444, P<10−5). Pairwise comparisons between each population pair (ignoring microhabitat stratification) yielded FST values of 0.0026 for Mk vs RM (P=0.009), 0.0036 for Mk vs SH (P=0.001) and 0.0022 for RM vs SH (P=0.069). FST values for microhabitats within each population are approximately fivefold larger: 0.0103 for Mk (P=0.883), 0.0102 for RM (P=0.0001) and 0.0103 for SH (P=0.773). Yet, these FST values for microhabitats are not significant, expect for RM, suggesting that there is large variation among SNPs such that the random permutation of individuals’ group assignments used to calculate P-values often capture similar large FST values.
SNP-specific FST values
Analyses using all SNPs together ignore evolutionary forces affecting one or a limited number of loci. To examine the SNP-specific differences among microhabitats, we employed three tests: SNP-specific FST and two common outlier tests (Figure 2). These tests are not independent because they rely on the same data (SNP-specific allele frequencies). Yet, they assess different aspects of the data with different assumptions. Significant FST values are based on a comparison of the FST value each SNP to the random distribution of FST values for that SNP generated by 1000 random permutations of individuals assigned to different microhabitats. This is a locus-specific test that does not consider the distribution of other SNPs (that could have a wide distribution with long tails; Storz, 2005). The two outlier tests, LOSITAN and Arlequin, are based on FDIST (Antao et al., 2008; Excoffier and Lischer, 2010) that defines significance relative to other similar SNPs. LOSITAN evaluates the relationship between FST and He (expected heterozygosity) in an island model, describing the expected distribution of FST values vs He under an island model of migration based on 50 000 simulations. This distribution is used to identify SNPs that have excessively high FST values compared with neutral expectations (Antao et al., 2008; Supplementary Figure 2). Such outlier SNPs are potentially evolving by natural selection. The distribution of P-value vs FST value for SNPs among microhabitats within a population using LOSITAN is provided in Supplementary Figure 3. Arlequin uses a hierarchical model, and coalescent simulations are used to get a null distribution and confidence intervals around the observed values and then tested to determine whether observed SNP-specific FST values can be considered as outliers conditioned on the globally observed FST value (Excoffier et al., 2009).
Outlier identification
We focused on SNPs that were significant in all three analyses (P<0.01 in each test (significant FST, LOSITAN and Arlequin); Figure 2), and where the joint probability of the three analyses had a FDR of ⩽1%, these were deemed outlier SNPs. The numbers of outlier SNPs among microhabitats for the three populations range from 63 to 110 (1.3 to 2.3% of SNPs; Table 4) with FST values that ranged from 0.08 to 0.31 (‘Outlier’, Figure 3a and Table 4). The means of outlier FST values among microhabitats are larger than the FST values among microhabitats determined using all 4741 SNPs (0.0102–0.0103), the average FST value when individuals are randomly assigned to one microhabitat (mean=−0.01±95% confidence interval (CI) 0.001) or for 4352 nonsignificant (mean=0.01±95% CI 0.001). These data demonstrate that significant, large FST values specific for a few hundred SNPs are unlikely to occur when individuals are randomly assigned to the three microhabitats (‘Randomized’, Figure 3a) and exceed the FST values for the 4352 SNPs that were not significant for any within-population comparison (‘Non-outlier’, Figure 3a). Thus, 1.3–2.3% of outlier SNPs have large microhabitat-specific FST values.
Distribution of FST values. (a) Density of FST values within each of the three replicate populations. Plotted are large significant outlier SNPs (blue), 4352 non-outlier SNPs (gold) and SNPs when population assignment is randomly permuted among microhabitats (red). (b) Density of outlier-SNP FST values within and among populations. Significant outlier SNP-specific FST values for within Rutgers University Marine Field Station (RUMFS, blue) and between RM and Stone Harbor (SH, gold) or Mantoloking (Mk, red).
There are 261 unique, significant, outlier SNPs among microhabitats. Four of these SNPs are significant outliers in two of the populations, but none are outliers among microhabitats in all three populations (Supplementary Table 1, ‘Population Significant’ column).
We calculated the LD for the 261 outlier SNPs to other SNPs within a 50-SNP window using the 15 259-SNP data set (Supplementary Figure 1c). There are 13 050 LD pairs, but only 1192 are significant. Of these 1192 significant LD pairs, most (1125, 94%) are only 100 bp apart. Thus, most outlier SNPs have few significant linkages to any other SNP >100 bp away (Supplementary Figure 1c).
Discriminant analysis of principle components
To provide insight into the differences among microhabitats, we employed DAPC (Jombart et al., 2010), a multivariate analysis that attempts to maximize the differences between groups while simultaneously minimizing the differences within groups. DAPC with the neutral SNPs (1636 SNPs) reveal large overlap among all nine microhabitats (Supplementary Figure 4a) and among microhabitats within a population (Supplementary Figures 4ai–iii). Using the 261 outlier SNPs found among microhabitats in each population, differences among microhabitats are seen among the 9 sampling locations (Supplementary Figure 4bi). This is most readily seen when plotting each population separately (Supplementary Figures 4bi–iii). Yet, among all 9 locations, microhabitats are not partitioned together: the first axis separates RM microhabitats, the second axis separates SH microhabitats and both axes separate Mk microhabitats. This most likely reflects the absence of shared outliers among the same microhabitats. It should not be surprising that SNPs with significant FST values provide strong DAPC (Figure 4b). Yet, these data show that a minority of SNPs suggests genetic divergence among microhabitats relative to a majority of SNPs.
Latitude vs FST values. Red points are FST values from using 11 microsatellites from Duvernell et al. (2008). Blue points are FST values from LDH-B allozyme data from Powers and Place (1978). Yellow points are FST values calculated among all 4741 SNPs. Black points are the FST values of outlier SNPs identified within each population among microhabitats.
Annotations and GO enrichment
The sequence tags for 265 outlier SNPs (Figure 2) that occur in any of the three populations all aligned to the Fundulus genome. Four of 265 outlier genes are shared between pairs of populations, yielding 261 unique outlier SNPs. For the 261 unique outlier SNPs among the three replicate populations (Figure 2), we determined the type (coding, 3′ noncoding, intergenic and so on), location and annotation (Supplementary Table 1). In all, 194 of the 261 unique outlier SNPs are in, or near, genic regions (Supplementary Table 1) with identified gene products (NCBI). Four outlier SNPs alter codons, and two of these are nonsynonymous (Rho GTPase and splicing factor U2AF 65 kD). However, most (134, 52%) of the unique outlier SNPs are intronic, and 67 (26%) are intergenic (between annotated genes and >5 kb away). Fifty-six (22%) outlier SNPs are upstream (41, 16%), downstream (14, 5%) or in untranslated regions (1), where upstream or downstream is within 5 kb of an annotated gene (Supplementary Table 1).
Whereas no outlier SNPs are shared among all three populations and only four of the same SNPs are shared among two populations (Supplementary Table 1, ‘Population Significant’ column), 11 outlier SNPs are shared among populations but are found (1) in the same gene at a different position, (2) in a duplicate gene or paralog or (3) among genes with similar annotations. One example is outlier SNPs in RM and SH populations that are 33 bp apart in the same intron of vav guanine nucleotide exchange factor 2 (signal transduction). Three sets of paralogs have outlier SNPs that affect all three replicate populations: (1) glutamate receptor (GRM4, GRM4’ and GRM5, where GRM4 and GRM4’ are two duplicate genes on different scaffolds), (2) serine/threonine kinases (STK10, STK32A, STK32c) and (3) zinc-finger proteins (ZNF19, ZNF19’ and ZNF507, where ZNF19 and ZNF19’ are two duplicate genes on different scaffolds). We note that there is no association between the scaffold number and shared chromosomes, but instead scaffolds are ranked by the size of the scaffold.
Of the outlier SNPs, 157 have human homologs recognized by PANTHER (Mi et al., 2016) and were used to define GO. These 157 human genes have nearly identical gene descriptions as those in the Fundulus genome (NCBI: GCA_000826765.1 Fundulus_heteroclitus-3.0.2). Gene ontologies for biological processes using 157 human homologs versus ~21 000 human proteins reveal enrichment for 23 protein classes. The most significant (P<10−5) was for 22 outlier SNPs associated with ‘intracellular signal transduction’ (IST). This GO category captures the glutamate receptor and serine/threonine kinase described above. With 22 outlier SNPs associated with IST, it is not surprising that there are shared outliers among all three replicate populations. Yet, examining subterms or GO ‘children’ of IST reveals more specific shared responses. The subterm within IST, ‘calcium-mediated signaling’ (enrichment P<0.03), has only three genes (VAV2, CAMK1D and KCNIP4) that have four SNPs shared across all three replicate populations. Similarly, within ‘cell communication’ is the subterm ‘neuron-neuron synaptic transmission’ with three genes (NNST, P<0.015). These three NNST genes have three outlier SNPs shared across all three populations. The NNST genes are the glutamate receptors (GRM4, GRM4’ and GRM5) that occur on three separate scaffolds (the two GRM4 are duplicate genes) and are outliers in RM, SK and Mk, respectively. The glutamate receptors are post synaptic, and we find it interesting that in presynaptic genes are four SNPs in ‘synaptic vesicle exocytosis gene’ (P<0.005) that occur predominately only in Mk (one SNP is shared with RM and Mk). Examining specific GO terms (<5 genes per term) for molecular function revealed two GO terms that were shared among all three replicate populations: ‘exoribonuclease activity’ (CNOT6l, CNOT8 XRN1) and ‘voltage-gated potassium channel activity’ (KCNIP4, KCNMA1 HCN1). Overall, based on restricted GO terms (<5 genes per term), 12 outlier SNPs are shared among all three replicate populations.
Discussion
Among microhabitats within a marsh estuary, F. heteroclitus share a common reproductive area in the high upper tidal zone, have few physical barriers to movement among microhabitats and are often transported by incoming and outgoing tides, creating what appears to most ecologists as a well-mixed population (Lotrich, 1975a; Kneib, 1978; Teo and Able, 2003b; Able et al., 2006, 2012). Thus, with large effective population size (Adams et al., 2006; Duvernell et al., 2008) and mobility among microhabitats (Able et al., 2012), there should be little neutral divergence among microhabitats. This view of Fundulus saltmarsh community is support by FST values using all 4741 SNPs where there are few significant differences among all nine collection sites (three populations each with three microhabitats, Table 3). In addition, a vast majority of SNPs have small FST values (average of ~0.01, Table 4 and Figure 3a). In general, non-outlier SNPs (SNPs that lack significant FST values among any microhabitat) are similar to microsatellites showing small changes among populations (Figure 4) and little difference among microhabitats (Figure 3a). Thus, there does not seem to be extensive isolation among microhabitats based on most of the SNPs.
Although there is little genetic divergence among microhabitats based on all SNPs (FST=0.01) or the average non-outlier SNPs (FST=−0.01, 95% CI −0.011 to −0.007, Table 4), there are 63 to 110 significant outlier SNPs (Figure 2) with large FST values among microhabitats within each of the 3 populations studied (Table 4, average FST=0.15, 95% CI 0.14–0.16). The FST values for outlier SNPs greatly exceed the FST values among populations for the same SNPs (Figure 3b) and for most non-outlier SNPs (Figure 3a). That is, FST values for outlier SNPs have a distribution that is unlike that of non-outlier SNPs or FST values among populations. The difference between the few hundred outlier SNPs versus the vast majority, we suggest, is related to ecological differences among microhabitats.
Saltmarsh estuaries have distinct microhabitats, including permanent ponds, tidal basins and intertidal creeks. These microhabitats are environmentally different and affect growth and survival. For example, ponds, which receive intermittent, nondaily, flushes of tidal water and experience high daily temperature maximum and low nightly dissolved oxygen, are more productive than basins or creeks (Teo and Able, 2003b; Able et al., 2006, 2012). In contrast, tidal basins have lower daily environmental fluxes in water temperature, salinity and dissolved oxygen (Halpin, 2000; Teo and Able, 2003a, b; Hunter et al., 2007). Intertidal creeks are dry at low tide and are then flooded by incoming tides. For ponds, although there is large annual influx, there is little immigration or emigration after April, suggesting many individuals spend a significant portion of their active season only in ponds (Able et al., 2012).
Although adult F. heteroclitus seem to flow in and out with the tidal cycles and reproduce in a common area, they have small home ranges and high site fidelity (Lotrich, 1975a; Skinner et al., 2005, 2012; Able et al., 2012). In a study of 1499 marked fish over 60 days, F. heteroclitus exhibited a 36 m home range, with the greatest distance moved being 375 m by just three fish; also, individuals had site fidelity (returning to the same side of a creek after release; Lotrich, 1975b). Similarly, in a separate study, 97% of tagged individuals were found within 200 m of their initial marking site over two seasons (Skinner et al., 2005), and stable isotopes indicate very few F. heteroclitus (3.4%) move >200 m in their lifetime (Skinner et al., 2012). For young of the year, 54% were recaptured within 5 m of the initial tagging site (Able et al., 2006). These data are supported by a remarkable mark–recapture study of >14 000 individuals over 17 months, where the authors concluded that despite physical connectivity of saltmarsh estuaries, there was almost complete fidelity to a single creek (Able et al., 2012). In addition, this research (Hunter et al., 2007; Able et al., 2012) demonstrated that ponds have resident fish with small emigration and immigration after April, many (75–95%) fish are recaptured in the same microhabitat and most movement (as measured by recapture) is from the basin into other microhabitats.
These data of microhabitat fidelity are supported by our SNP data. When considering all 4741 SNPs, the AMOVA analysis revealed that 10 times as much variation is explained by microhabitats than between populations along the NJ coast (Table 2). Among microhabitats within a population, FST values (0.007) based on all 4741 SNPs are much larger than the FST values among populations along the NJ coast (0.0026–0.0036). Thus, microhabitats within a saltmarsh estuary have population structure that exceeds that among distant populations (Mk, RM and SH). More important are the large FST values among microhabitats for the outlier SNPs. The large FST values among microhabitats within a population for outlier SNPs and the observation that these FST values exceed those among geographically distant populations are indicative of fine-scale genetic structure. The evolutionary importance of this observation depends on two points: (1) whether outlier FST values are statistical errors (type I errors) or (2) whether neutral evolutionary processes are likely to explain the large outlier FST values.
Significance of outlier SNPs
We defined significant outlier SNPs as those significant in all three analyses (P<0.01 in each test (significant FST, LOSITAN and Arlequin); Figure 2) and where the joint probability of the three analyses had a FDR of ⩽1%. The FST values for these 63–110 outlier SNPs (1.3–2.3% of SNPs; Table 4) range from 0.08 to 0.31 and exceed the distribution of the vast majority of nonsignificant SNPs (Figure 3a and Table 4). These data demonstrate that the outlier SNPs have significant, large FST values that are not seen for most SNPs; nor are they seen when individuals are randomly assigned to one of three microhabitats (‘random’, Figure 3a). That is, when individuals are randomly assigned to a microhabitat in 1000 permutations, the average FST value is less than zero (Table 4), and only 0.4% of >13 million random FST values exceed the lower 95% CI of the outlier SNPs (Table 4). Yet, the true neutral FST value distribution could have a wide distribution with long tails that includes large FST values (Lotterhos and Whitlock, 2015; Whitlock and Lotterhos, 2015).
Although outlier SNPs tests are designed to identify adaptive divergence (Beaumont and Balding, 2004; Antao et al., 2008; Excoffier et al., 2009), they are likely to have false positives (Lotterhos and Whitlock, 2014), and the significant outliers instead reflect neutral processes because demography and migration can affect FST value variance among SNPs (Lotterhos and Whitlock, 2014). The false positive rate is related to the population’s demography and the specific demographic model of the outlier tests (Lotterhos and Whitlock, 2014). If comparisons are among populations that are isolated by distance or match an island model used in FDIST2 as implemented by LOSITAN (Antao et al., 2008), false positive rates are low (<5%) (Lotterhos and Whitlock, 2014). However, if the population reflects expansion from one or two refugia, then false discovery rates become unacceptably high (Lotterhos and Whitlock, 2014). For saltmarsh microhabitats, there is little reason to believe there is any population structure, and this is supported by 4352 (91%) SNPs that have very small and nonsignificant FST values within a population (mean=0.009–0.010; Table 4 and Figure 3a). With no expected demographic structure and little support for population structure for most SNPs, the magnitude of errors for these outlier tests is unclear. However, the observations that >190 SNPs in each population (Figure 2) have significant FST values and both outlier tests are significant suggest that there are hundreds of SNPs with exceptionally high FST values (Figure 3a). Thus, we are suggesting that many of the outlier SNPs have unlikely large FST values that are true positives.
The statistical approach presented here differs from similar studies demonstrating unexpected genetic divergences (Hess et al., 2013; Bergland et al., 2014) in that we rely on three separate analyses. Whereas a single test is likely to suffer from false positives, three separate tests are unlikely to simultaneously be falsely positive (Lotterhos and Whitlock, 2014, 2015). Instead, our conservative approach is more likely to suffer from false negatives or large type II errors (falsely accepting the null hypothesis of no differences among groups (de Villemereuil et al., 2014; Lotterhos and Whitlock, 2014)). We likely are missing many outlier SNPs because of our approach.
We conclude that some of our outlier FST values may be false positives, and an equal or greater number of SNPs may be false negatives (de Villemereuil et al., 2014; Lotterhos and Whitlock, 2014). Yet, the identification of hundreds of SNPs with large significant FST values found in three separate analyses suggested evolutionary forces effecting a significant allele frequency change.
Genomic differentiation among microhabitats
We suggested that the outlier SNPs are related to ecologically relevant differences in temperature, salinity, frequency of hypoxic events and productivity among microhabitats (Teo and Able, 2003b; Able et al., 2006, 2012). What are the evolutionary forces that would create these large significant FST values among microhabitats? Neutral mutation–drift process could affect FST values, where a rare allele rises to appreciable frequency. Yet, the outlier SNPs do not have rare or low frequency alleles: for outlier SNPs, the MAFs within a single microhabitat are on average 0.376, and range from 0.072 to 0.912 (where MAF is defined across all samples). The significantly large FST values arise because these MAFs may have much higher frequencies within a specific microhabitat. This, plus the large effective population size of F. heteroclitus (Duvernell et al., 2008), makes it unlikely that neutral mutation–drift processes are responsible for the large FST values among outlier SNPs. We also suggest that mutation drift is unlikely to be responsible for the outlier-SNP FST values among microhabitats because it would take many generations with little connectivity for a mutation to rise to the MAF observed here, and the natural history of F. heteroclitus suggests large generational connectivity among microhabitats. That is, because F. heteroclitus breed in a common area and live for 3 years in the field (Valiela et al., 1977), each generation should be dispersed among microhabitats unless individuals select microhabitat based on their genotype.
It is possible that migrants from populations with significant allele frequency differences at some SNPs could inflate FST values within a population if these migrants ended up in one microhabitat. Similar to allele surfing (Excoffier and Ray, 2008), a few invaders could cause large FST values for some loci by chance. To examine this, we compared the FST values for the 63 outlier SNPs within RM with the FST values for the same 63 SNPs in populations north (Mk) or south (SH) of RM. The FST values for outlier SNPs within a population among microhabitats exceed the FST values between populations (Figure 3b). The average FST values (0.15) for the outliers within RM are much greater than the FST values for the same outlier-SNP FST values between populations north (0.01) or south (0.009) of RM (Figure 3b). Because outlier-SNP FST values between microhabitats are much larger than between populations, it seems unlikely that random migration from a different population to a specific microhabitat would be responsible for the difference among microhabitats. The outlier-SNP FST values among microhabitats are also greater than values reported previously: the FST values based on microsatellites (Duvernell et al., 2008) or the LDH-B locus that is most likely evolving by natural selection (Powers et al., 1991; Figure 4). Among populations, the microsatellites (Duvernell et al., 2008) and the non-outlier-SNP FST values are similar and low, suggesting small neutral divergence among populations. Not only are outlier-SNP FST values calculated among microhabitats larger than the differences calculated among populations, but they also exceed previously published data for neutral (Duvernell et al., 2008) and adaptive (Powers et al., 1991) divergences among populations. Thus, the data do not support migration from distant populations into specific microhabitats.
We suggest that neutral processes including nonrandom migration are an unlikely cause of outlier-SNP FST values, yet we need to point out that nearly all outlier SNPs were unique to their population. Only four outlier SNPs are significant between any two populations, and no outlier SNP was an outlier in all three populations. To examine whether our conservative filtering to 4761 SNPs affects how many significant outlier SNPs are shared among the three replicate populations, we applied LOSITAN outlier analyses to the 15 259 SNPs (this includes SNPs in LD and with no minimum allele frequency cutoff). None of the significant outlier SNPs using 15 259 is an outlier in all 3 replicate populations, and only 24 of 1022 (2%) outlier SNPs were shared between any 2 replicate populations. Thus, few of the same SNPs have a shared response among the three replicate populations (Mk, RM and SH) even when considering the larger 15K-SNP set.
A greater percentage of outlier SNPs shared among populations and among microhabitats would have strengthened our analyses (Figure 2). The lack of shared outlier SNPs among the three replicate populations has three potential explanations. (1) The absence of shared outlier SNPs suggests that the test for outlier-SNP FST values underestimates the length and complexity of the neutral FST value distribution. Thus, our significant outlier SNPs represent random neutral divergences that we would not expect to be shared. (2) The total length of the 4741 tag sequences used to identify SNPs is only 0.04% of the genome (Reid et al., 2015). With so little genome coverage, it is possible that we missed many more outlier SNPs shared among the three replicate populations. (3) There may be many different loci where a relatively small allele frequency change can affect an evolutionarily important phenotypic change (Przeworski et al., 2005; Pritchard et al., 2010). This hypothesis of polygenic selection from large standing genetic variation would provide many potential solutions involving a subset of many different genes, and thus it is unlikely that selection would favor the same SNPs in all populations.
The first potential explanation suggests the possibility that neutral FST values have a very wide distribution. Yet, given the data on the genetic divergence within and among populations it unlikely that neutral processes are responsible for these large FST values. The distributions of FST values found among populations or among the random permutations do not include the large FST values found with the significant outlier SNPs, and thus it seems unlikely that these outlier-SNP FST values occur by random or other neutral demographic processes.
The second potential explanation proposes that many common outlier SNPs were missed. This could happen because so little of the genome was examined, but our results can be compared with 384 Fundulus whole-genome sequences defined in four pairs of populations subject to recent anthropogenic pollution (Reid et al., 2016). The whole-genome adaptive changes identified among polluted populations rarely involve the same exact change; instead, adaptive changes often involve a variety of changes in the same genes, or genes in the same pathway (Reid et al., 2016). The annotation and gene ontologies for the outlier SNPs presented here can be similarly interpreted—all three populations share outlier SNPs in three glutamate receptors, four calcium signaling proteins, three serine/threonine kinases or three zinc-finger proteins. Thus, data on a small percentage of the genome are not fundamentally different than whole-genome sequence analyses.
The third explanation, polygenic selection from standing genetic variation, has support from our data: among 15 259 SNPs there are 8180 (12% of all possible) significant LDs and 93% of these are <100 bp apart. For the 261 outlier SNPs, there are 1192 significant LDs with 15 259 other SNPs, and all but 67 or 94% of these are <100 bp apart. In the classic model of evolution, where new mutations quickly go to fixation when affected by nature selection, outlier SNPs should be associated with large linkage blocks. Clearly, this is not what we find, nor would we expect to if the microhabitats within a population are inundated with young of year every spring. Instead, the short distances among all SNPs and among 94% of outlier SNPs with significant LD is indicative of long-term standing genetic variation.
This explanation, that the lack of long LD blocks reflects outlier SNPs affecting polygenic traits and where many different genes provide similar solutions to resolve natural selection for a phenotypic trait, is supported by annotations (Supplementary Table 1). Different outlier SNPs among the three replicate populations occur in the same gene, but at different positions in the same gene or in paralogs: metabotropic glutamate receptor 4 and 5, three serine/threonine kinases (STK10, STK32A and STK32c) and three zinc-finger proteins (ZNF19, ZNF19’ and ZNF507). Similarly, all three populations share specific GO terms with 5 genes: (1) three ‘calcium-mediated signaling’ genes (enrichment P<0.03, VAV2, CAMK1D and KCNIP4), (2) three ‘neuron-neuron synaptic transmission’ genes (P<0.015, GRM4, GRM4’ and GRM5), (3) three ‘RNA exoribonuclease’ (P<0.005, CNOT6, CNOT8 and XNR1) and (4) ‘three voltage-gated potassium channel activity’ (KCNIP4, KCNMA1 HCN1). The observation that all three populations share two sets of paralog genes (STK, ZNF) or four finite GO functions suggests that evolutionary forces are acting on similar biological processes among microhabitats across all three populations.
We suggest that the unlikely, large FST values with little long-distance LD among genes of similar functions are a hallmark of natural selection (Beaumont and Balding, 2004; Excoffier et al., 2009) and that natural selection is acting on each generation effecting small but significant allele frequency changes in some of many potential genes that respond to natural selection. That is, there is polygenic selection, and only a subset of possible adaptive genes respond to natural selection in each population (Przeworski et al., 2005; Pritchard et al., 2010)
We conclude that although microhabitats are well connected within a saltmarsh estuary, natural selection is effecting allele frequency changes and is responsible for the fine-scale geographic structure. This conclusion must be tentative because of its repercussions for previous fieldwork on Fundulus and, to be frank, the highly unlikely nature of adaptive divergence affecting demes inhabiting microhabitats. What is required are temporal studies of pond- and basin-resident fishes to determine whether microhabitat residents with specific genotypes are more likely to survive the summer. These studies would reveal whether a common set of genes are selected in different microhabitats and whether the genes being selected are adaptively significant in one, several or all microhabitats.
Evolution among microhabitats
In 1966, Lewontin and Hubby (1966) provided data on 18 proteins and concluded that much genetic variation exists within a species, but they could not resolve the evolutionary forces effecting this variation. After 50 years, it is not clear that we have resolved this dilemma (Ohta, 1992; Kreitman, 1996; Charlesworth, 2015; Henn et al., 2015). We demonstrate here that there are 63–110 SNPs with large and unlikely FST values (~0.15, ⩽1% FDR) among microhabitats in the three NJ populations. We argue above that migration, demography or other neutral processes are not responsible, and thus suggest that natural selection is changing allele frequencies. The observation that there is greater variation among microhabitats than among populations (Table 2) is similar to the classic indication of selection: greater variation within a species than between species (Slatkin, 1987; Kreitman, 1996). If natural selection were responsible, given the natural history of F. heteroclitus, it would require annual selection of specific genotypes in ponds, creeks and basins. Because ponds are environmentally different, adaptation to this environment seems possible. Our reluctance to invoke selection is that this selection would have to occur in the presence of large annual immigrations of new individuals (Able et al., 2006, 2012). This would seem to create a substantial genetic load (Haldane, 1957). However, recent investigations of genetic load in human populations suggests that contemporary rates of human migration do not seem to substantially increase the number or occurrence of deleterious alleles (Henn et al., 2015). Charlesworth (2013) addresses this point directly, suggesting that large perceived genetic load can be mitigated by weak stabilizing selection and soft selection and reiterates that there is rarely an ‘optimum’ phenotype in each environment against which other organisms are competing. Rather, most individuals’ fitness is relative to the fitness of other individuals with whom they compete. Given the relatively small home ranges of F. heteroclitus (<200 m), this could contribute to the structure and genotypic diversity detected between the saltmarsh microhabitats.
Conclusion
All the evidence suggests that saltmarsh estuary populations of F. heteroclitus are not panmictic; rather, they exhibit a complex genetic landscape that creates microhabitat-specific demes with significant allele frequency differences at hundreds of loci, many of which share common biological functions. These data are similar to the large seasonal variation in thousands of SNPs in a single D. melanogaster population (Bergland et al., 2014). Despite the absence of physical or physiological barriers to migration, our evidence suggests that the individuals in different microhabitats within the same saltmarsh estuary belong to genetically divergent groups. The fine-scale (<200 m) population structure that this study reveals has yet to be documented in another vertebrate species, likely because it is so unexpected. This leads us to conclude: directional selection affects many SNPs, and these SNPs are different from population to population.
Data archiving
Three tab-delimited files are archived at Dryad (http://dx.doi.org/10.5061/dryad.mc04d). The first is the raw hapmap file with all called SNPs (338 325) and all individuals (n=215). The second is the 15 259 SNP file with 202 individuals used for to calculate distances among SNPs and significant LD. The third is the filtered hapmap with 4741 SNPs from 202 individuals that was used in all the analyses presented here.
References
Able KW, Hagan SM, Brown SA . (2006). Habitat use, movement, and growth of young-of-the-year Fundulus spp. in southern New Jersey salt marshes: comparisons based on tag/recapture. J Exp Mar Biol Ecol 335: 177–187.
Able KW, Vivian DN, Petruzzelli G, Hagan SM . (2012). Connectivity among salt marsh subhabitats: residency and movements of the mummichog (Fundulus heteroclitus). Estuar Coast 35: 743–753.
Adams SM, Lindmeier JB, Duvernell DD . (2006). Microsatellite analysis of the phylogeography, Pleistocene history and secondary contact hypotheses for the killifish, Fundulus heteroclitus. Mol Ecol 15: 1109–1123.
Antao T, Lopes A, Lopes RJ, Beja-Pereira A, Luikart G . (2008). LOSITAN: a workbench to detect molecular adaptation based on a F(st)-outlier method. BMC Bioinformatics 9: 323.
Baris TZ, Wagner DN, Dayan DI, Du X, Blier PU, Pichaud N et al. (2016). Evolve genetic and phenotypic differences due to mitochondrial-nuclear interactions. PLoS Genet. (In press).
Beaumont MA, Balding DJ . (2004). Identifying adaptive genetic divergence among populations from genome scans. Mol Ecol 13: 969–980.
Beaumont MA, Nichols RA . (1996). Evaluating loci for use in the genetic analysis of population structure. Proc R Soc B Biol Sci 263: 1619–1626.
Benjamini Y, Hochberg Y . (1995). Controlling the False Discovery Rate - a Practical and Powerful Approach to Multiple Testing. J R Stat Soc Series B Methodol 57: 289–300.
Bergland AO, Behrman EL, O’Brien KR, Schmidt PS, Petrov DA . (2014). Genomic evidence of rapid and stable adaptive oscillations over seasonal time scales in Drosophila. PLoS Genet 10: e1004775.
Bradbury PJ, Zhang Z, Kroon DE, Casstevens TM, Ramdoss Y, Buckler ES . (2007). TASSEL: software for association mapping of complex traits in diverse samples. Bioinformatics 23: 2633–2635.
Charlesworth B . (2013). Why we are not dead one hundred times over. Evolution 67: 3354–3361.
Charlesworth B . (2015). Causes of natural variation in fitness: evidence from studies of Drosophila populations. Proc Nat Acad Sci USA 112: 1662–1669.
Corander J, Majander KK, Cheng L, Merila J . (2013). High degree of cryptic population differentiation in the Baltic Sea herring Clupea harengus. Mol Ecol 22: 2931–2940.
Crawford DL, Oleksiak MF . (2016). Ecological population genomics in the marine environment. Brief Funct Genomics 15: 342–351.
Crow JF . (1958). Some possibilities for measuring selection intensities in man. Hum Biol 30: 1–13.
Danecek P, Auton A, Abecasis G, Albers CA, Banks E, DePristo MA et al. (2011). The variant call format and VCFtools. Bioinformatics 27: 2156–2158.
de Villemereuil P, Frichot E, Bazin E, Francois O, Gaggiotti OE . (2014). Genome scan methods against more complex models: when and how much should we trust them? Mol Ecol 23: 2006–2019.
Duvernell DD, Lindmeier JB, Faust KE, Whitehead A . (2008). Relative influences of historical and contemporary forces shaping the distribution of genetic variation in the Atlantic killifish, Fundulus heteroclitus. Mol Ecol 17: 1344–1360.
Elshire RJ, Glaubitz JC, Sun Q, Poland JA, Kawamoto K, Buckler ES et al. (2011). A robust, simple genotyping-by-sequencing (GBS) approach for high diversity species. PLoS One 6: e19379.
Excoffier L, Hofer T, Foll M . (2009). Detecting loci under selection in a hierarchically structured population. Heredity 103: 285–298.
Excoffier L, Lischer HEL . (2010). Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol Ecol Resour 10: 564–567.
Excoffier L, Ray N . (2008). Surfing during population expansions promotes genetic revolutions and structuration. Trends Ecol Evol 23: 347–351.
Excoffier L, Smouse PE, Quattro JM . (1992). Analysis of molecular variance inferred form metric distances among DNA haplotypes—application to human mitochondrial-DNA restriction data. Genetics 131: 479–491.
Haldane JBS . (1957). The cost of natural selection. J Genet 55: 511–524.
Halpin PM . (2000). Habitat use by an intertidal salt-marsh fish: trade-offs between predation and growth. Mar Ecol Prog Ser 198: 203–214.
Henn BM, Botigue LR, Bustamante CD, Clark AG, Gravel S . (2015). Estimating the mutation load in human genomes. Nat Rev Genet 16: 333–343.
Hess JE, Campbell NR, Close DA, Docker MF, Narum SR . (2013). Population genomics of Pacific lamprey: adaptive variation in a highly dispersive species. Mol Ecol 22: 2898–2916.
Huang W, Massouras A, Inoue Y, Peiffer J, Ramia M, Tarone AM et al. (2014). Natural variation in genome architecture among 205 Drosophila melanogaster Genetic Reference Panel lines. Genome Res 24: 1193–1208.
Hunter KL, Fox MG, Able KW . (2007). Habitat influences on reproductive allocation and growth of the mummichog (Fundulus heteroclitus) in a coastal salt marsh. Mar Biol 151: 617–627.
Ivanova NV, Dewaard JR, Hebert PDN . (2006). An inexpensive, automation-friendly protocol for recovering high-quality DNA. Mol Ecol Notes 6: 998–1002.
Jombart T . (2008). adegenet: a R package for the multivariate analysis of genetic markers. Bioinformatics 24: 1403–1405.
Jombart T, Ahmed I . (2011). adegenet 1.3-1: new tools for the analysis of genome-wide SNP data. Bioinformatics 27: 3070–3071.
Jombart T, Devillard S, Balloux F . (2010). Discriminant analysis of principal components: a new method for the analysis of genetically structured populations. BMC Genet 11: 94.
Kimura M . (1968). Evolutionary rate at the molecular level. Nature 217: 624–626.
Kneib RT . (1978). Habitat, diet, reproduction and growth of the spotfin killifish, Fundulus luciae, from a North Carolina salt marsh. Copeia 1978: 164–168.
Kneib RT . (1986). The role of Fundulus heteroclitus in salt-marsh trophic dynamics. Am Zool 26: 259–269.
Kreitman M . (1996). The neutral theory is dead. Long live the neutral theory. Bioessays 18: 678–683.
Lewontin RC . (1964). The interaction of selection and linkage. I. General considerations; heterotic models. Genetics 49: 49–67.
Lewontin RC, Hubby JL . (1966). A molecular approach to the study of genic heterozygosity in natural populations. II. Amount of variation and degree of heterozygosity in natural populations of Drosophila Pseudoobscura. Genetics 54: 595–609.
Li H, Durbin R . (2009). Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25: 1754–1760.
Lind EE, Grahn M . (2011). Directional genetic selection by pulp mill effluent on multiple natural populations of three-spined stickleback (Gasterosteus aculeatus). Ecotoxicology 20: 503–512.
Lotrich VA . (1975a). Summer home range and movements of Fundulus heteroclitus (Pisces-Cyprinodontidae) in a tidal creek. Ecology 56: 191–198.
Lotrich VA . (1975b). Summer home range and movements of Fundulus heteroclitus (Pisces: Cyprinodotidae) in tidal creek. Ecology 56: 191–198.
Lotterhos KE, Whitlock MC . (2014). Evaluation of demographic history and neutral parameterization on the performance of FST outlier tests. Mol Ecol 23: 2178–2192.
Lotterhos KE, Whitlock MC . (2015). The relative power of genome scans to detect local adaptation depends on sampling design and statistical method. Mol Ecol 24: 1031–1046.
Mace ES, Phong DT, Upadhyaya HD, Chandra S, Crouch JH . (2006). SSR analysis of cultivated groundnut (Arachis hypogaea L.) germplasm resistant to rust and late leaf spot diseases. Euphytica 152: 317–330.
Mackay TF, Richards S, Stone EA, Barbadilla A, Ayroles JF, Zhu D et al. (2012). The Drosophila melanogaster Genetic Reference Panel. Nature 482: 173–178.
Messer PW, Petrov DA . (2013). Population genomics of rapid adaptation by soft selective sweeps. Trends Ecol Evol 28: 659–669.
Mi H, Poudel S, Muruganujan A, Casagrande JT, Thomas PD . (2016). PANTHER version 10: expanded protein families and functions, and analysis tools. Nucleic Acids Res 44: D336–D342.
Newman HH . (1907). Spawning behavior and sexual dimorphism in Fundulus heteroclitus and allied fish. Biol Bull 12: 314–348.
Nielsen EE, Hemmer-Hansen J, Poulsen NA, Loeschcke V, Moen T, Johansen T et al. (2009). Genomic signatures of local directional selection in a high gene flow marine organism; the Atlantic cod (Gadus morhua). BMC Evol Biol 9: 276.
Nunez JC, Seale TP, Fraser MA, Burton TL, Fortson TN, Hoover D et al. (2015). Population Genomics of the Euryhaline Teleost Poecilia latipinna. PLoS One 10: e0137077.
Ohta T . (1992). The nearly neutral theory of molecular evolution. Annu Rev Ecol Syst 23: 263–286.
Otto SP, Whitlock MC . (1997). The probability of fixation in populations of changing size. Genetics 146: 723–733.
Paradis E, Claude J, Strimmer K . (2004). APE: analyses of phylogenetics and evolution in R language. Bioinformatics 20: 289–290.
Powers DA, Lauerman T, Crawford D, Dimichele L . (1991). Genetic mechanisms for adapting to a changing environment. Annu Rev Genet 25: 629–659.
Powers DA, Place AR . (1978). Biochemical genetics of Fundulus heterolitus (L.). I. Temporal and spatial variation in gene frequencies of Ldh-B, Mdh-A, Gpi-B, and Pgm-A. Biochem Genet 16: 593–607.
Pritchard JK, Pickrell JK, Coop G . (2010). The genetics of human adaptation: hard sweeps, soft sweeps, and polygenic adaptation. Curr Biol 20: R208–R215.
Przeworski M, Coop G, Wall JD . (2005). The signature of positive selection on standing genetic variation. Evolution 59: 2312–2323.
Reid NM, Jackson CE, Gilbert D, Minx P, Montague MJ, Hampton TH et al. (2015). The Atlantic killifish (Fundulus heteroclitus) genome and the landscape of genome variation within a population. Biol Evol, (In Press).
Reid NM, Proestou DA, Clark BW, Warren WC, Colbourne JK, Shaw JR et al. (2016). The genomic landscape of rapid repeated evolutionary adaptation to toxic pollution in wild fish. Science 354: 1305–1308.
Skinner MA, Courtenay SC, Parker WR, Curry RA . (2005). Site fidelity of mummichogs (Fundulus heteroclitus) in an Atlantic Canadian estuary. Water Qual Res J Can 40: 288–298.
Skinner MA, Courtenay SC, Parker WR, Curry RA . (2012). Stable isotopic assessment of site fidelity of mummichogs, Fundulus heteroclitus, exposed to multiple anthropogenic inputs. Environ Biol Fishes 94: 695–706.
Slatkin M . (1987). Gene flow and the geographic structure of natural populations. Science 236: 787–792.
Storz JF . (2005). Using genome scans of DNA polymorphism to infer adaptive population divergence. Mol Ecol 14: 671–688.
Teo SLH, Able KW . (2003a). Growth and production of the mummichog (Fundulus heteroclitus) in a restored salt marsh. Estuaries 26: 51–63.
Teo SLH, Able KW . (2003b). Habitat use and movement of the mummichog (Fundulus heteroclitus) in a restored salt marsh. Estuaries 26: 720–730.
Turelli M, Barton NH . (2004). Polygenic variation maintained by balancing selection: Pleiotropy, sex-dependent allelic effects and GxE interactions. Genetics 166: 1053–1079.
Valiela I, Wright JE, Teal JM, Volkmann SB . (1977). Growth, production and energy transformations in the salt-marsh killifish Fundulus heteroclitus. Mar Biol 40: 135–144.
Weir BS, Cockerham CC . (1984). Estimating F-statistics for the analysis of population-structure. Evolution 38: 1358–1370.
Whitlock MC, Lotterhos KE . (2015). Reliable detection of loci responsible for local adaptation: inference of a null model through trimming the distribution of F-ST. Am Nat 186: S24–S36.
Williams GC . (1966) Adaptation and Natural Selection: A Critique of Some Current Evolutionary Thought. Princeton University Press: Princeton, NJ.
Yeaman S . (2015). Local adaptation by alleles of small effect. Am Nat 186: S74–S89.
Acknowledgements
We thank Rutgers University Marine Field Station, Roland Hagan, Kaycee Coleman and Ken Able for support during sample collection and lively discussion on evolutionary ecology. This research is supported by grant from NSF IOS 1147042 and NSF MCB 1158241. These analyses of these data were greatly enhanced by Fundulus heteroclitus genome supported by NSF DEB-1265282.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on Heredity website
Rights and permissions
About this article
Cite this article
Wagner, D., Baris, T., Dayan, D. et al. Fine-scale genetic structure due to adaptive divergence among microhabitats. Heredity 118, 594–604 (2017). https://doi.org/10.1038/hdy.2017.6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/hdy.2017.6
This article is cited by
-
Contamination acts as a genotype-dependent barrier to gene flow, causing genetic erosion and fine-grained population subdivision in Mussels from the Strait of Istanbul
Ecotoxicology (2024)
-
Transcriptomic analysis provides insights into molecular mechanisms of thermal physiology
BMC Genomics (2022)
-
Divergence of a genomic island leads to the evolution of melanization in a halophyte root fungus
The ISME Journal (2021)
-
Phenotypic differentiation in a resilient dung beetle species induced by forest conversion into cattle pastures
Evolutionary Ecology (2019)
-
Complex signatures of genomic variation of two non-model marine species in a homogeneous environment
BMC Genomics (2018)