Abstract
Ommochromes are major insect pigments involved in coloration of compound eyes, eggs, epidermis and wings. In the silkworm Bombyx mori, adult compound eyes and eggs contain a mixture of the ommochrome pigments such as ommin and xanthommatin. Here, we identified the gene involved in ommochrome biosynthesis by positional cloning of B. mori egg and eye color mutant pink-eyed white egg (pe). The recessive homozygote of pe has bright red eyes and white or pale pink eggs instead of a normal dark coloration due to the decrease of dark ommochrome pigments. By genetic linkage analysis, we narrowed down the pe-linked region to ~258 kb, containing 17 predicted genes. RNA sequencing analyses showed that the expression of one candidate gene, the ortholog of Drosophila haem peroxidase cardinal, coincided with egg pigmentation timing, similar to other ommochrome-related genes such as Bm-scarlet and Bm-re. In two pe strains, a common missense mutation was found within a conserved motif of B. mori cardinal homolog (Bm-cardinal). RNA interference-mediated knockdown and transcription activator-like effector nuclease (TALEN)-mediated knockout of the Bm-cardinal gene produced the same phenotype as pe in terms of egg, adult eye and larval epidermis coloration. A complementation test of the pe mutant with the TALEN-mediated Bm-cardinal-deficient strain showed that the mutant phenotype could not be rescued, indicating that Bm-cardinal is responsible for pe. Moreover, knockdown of the cardinal homolog in Tribolium castaneum also induced red compound eyes. Our results indicate that cardinal plays a major role in ommochrome synthesis of holometabolous insects.
Similar content being viewed by others
Introduction
Ommochromes represent a major group of insect pigments and are divided into two subgroups, ommins and ommatins, depending on their liability to alkaline conditions (Butenandt et al., 1958; Linzen, 1974). Ommins are stable under alkali conditions, a compound with a high degree of polymerization, and function as screening pigments in the eyes of most insects except for higher dipterans. Ommins are also found in the eyes of many arthropod and cephalopod species, and in the epidermis of the cricket Gryllus bimaculatus, the stick insect Carausius morosus and the mantis Mantis religiosa (Linzen, 1974). Ommatins are unstable under alkali conditions, are of rather low molecular weight and are found in epidermis, wings, excreta and eyes (Linzen, 1974; Nijhout, 1997). Xanthommatin is a common ommatin pigment found in the eyes of most insects. Xanthommatin has two forms, an oxidized form and a reduced form, that are associated with brown/red color differences of Heliconius butterfly wings (Gilbert et al., 1988) and yellow/red body color change of red dragonflies (Futahashi et al., 2012).
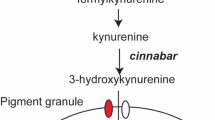
The genes involved in ommochrome biosynthesis have been identified mostly by genetic analyses of Drosophila eye color mutants. The primary ommochrome precursor is tryptophan that is converted into 3-hydroxykynurenine by three enzymes encoded by vermilion, kynurenine formamidase and cinnabar (Searles and Voelker, 1986; Searles et al., 1990; Warren et al., 1996). Subsequently, 3-hydroxykynurenine is incorporated into pigment granules by the heterodimeric ABC (ATP-binding cassette) transporters encoded by scarlet and white (Tearle et al., 1989; Pepling and Mount, 1990). However, the molecular mechanisms involved in the conversion of 3-hydroxykynurenine into ommochrome pigments are largely unknown, and this may be because of the lack of ommin pigments in Drosophila melanogaster (Linzen, 1974).
It has been reported that the eggs and adult compound eyes of the silkworm Bombyx mori contain a combination of the ommochrome pigments, ommin and xanthommatin (Kawase and Aruga, 1954; Koga and Osanai, 1967; Linzen, 1974; Sawada et al., 2000). Many Bombyx egg and eye color mutants have been preserved and used to study insect genetics such as homologous recombination (Ariga and Murakami, 1975; Ariga and Murakami, 1976) and parthenogenetic induction (Hirokawa, 1993; Hirokawa, 1996). B. mori egg and eye color mutants w-1, w-2 and w-3 have been reported to correspond to Drosophila eye color mutants cinnabar, scarlet and white, respectively, indicating that genes involved in the early steps of the ommochrome synthesis pathway are conserved between the two species (Quan et al., 2007; Komoto et al., 2009; Tatematsu et al., 2011). In contrast, some B. mori egg color mutants could not be explained by simple adaptation of Drosophila eye color mutants. We have reported that the gene responsible for B. mori red egg (re) mutant is a novel member of a major facilitator superfamily transporter that is conserved in most insects but is lost in Drosophila (Osanai-Futahashi et al., 2012). The re mutant is deficient in ommin synthesis, suggesting that the loss of the re gene ortholog in higher Diptera coincides with the lack of ommins in their eyes (Linzen, 1974). Owing to them having a more complex ommochrome composition than Drosophila and still unrevealed ommochrome mutants, B. mori is suitable for investigating the final steps of ommochrome biosynthesis.
Pink-eyed white egg (pe) is a recessive egg and eye color mutant of B. mori. The recessive homozygote of pe generally has white eggs and pink eyes, a brighter shade than those of the re mutant (Figure 1). Notably, the non-diapausing eggs of the pe mutant turn pink after several weeks of preservation at low temperature. The pe locus has been mapped to linkage group 5, the same as the re locus. In the pe mutants, the level of 3-hydroxykynurenine, the ommochrome precursor, accumulates compared with wild type, suggesting that pe mutants have defects in the conversion of 3-hydroxykynurenine to final ommochromes (Inagami, 1954). It has been suggested that pe functions downstream of w-2 (scarlet) and upstream of re from genetic studies (Uda, 1932).
Wild-type and pe mutant of the silkworm B. mori. Eggs (top), adult compound eyes (middle) and eyespot markings of final-instar larva (bottom) of wild-type (+pe/+pe, left) and pee20 mutant (pe/pe, right) are shown. Around the eyespot markings, red markings are found in the Striped (pS) strain (+pe/+pe; pS/pS, bottom left) but not in double mutants of pS and pe (pe/pe; pS/pS, bottom right).
Here, we identify the gene corresponding to the pe locus by positional cloning. We report a missense mutation in a gene encoding peroxidase orthologous to the cardinal gene of D. melanogaster (Harris et al., 2011), and reproduce the pe phenotype by RNA interference (RNAi) gene knockdown and transcription activator-like effector nuclease (TALEN)-mediated gene knockout. We also conducted a complementation test by crossing gene knockout individuals with pe mutants, and found that these were unable to rescue the pe phenotype, demonstrating that the B. mori cardinal homolog is the gene responsible for the pe phenotype. We also confirmed the decrease of red/purple pigments (supposed ommin), decarboxylated xanthommatin and xanthommatin in pe mutants by thin-layer chromatography (TLC) and high-pressure liquid chromatography–mass spectrometry (HPLC-MS). Instead, the ommochrome precursor 3-hydroxykynurenine and uncharacterized pigments seem to accumulate in pe mutants. Moreover, in the red flour beetle Tribolium castaneum, knockdown of the cardinal homolog also induced the red eye phenotype, suggesting that the cardinal gene has a major role in biosynthesis of normal ommochrome pigments, at least in holometabolous insects.
Materials and methods
Silkworm and Tribolium strains
We used two pe strains, pe re911 and pee20, for this study. The pee20 mutant strain was provided from the silkworm stock center of Kyushu University (Fukuoka, Japan) supported by the National BioResource Project (Banno et al., 2010). The wild-type silkworm strain p50T was maintained in the Transgenic Silkworm Research Unit, National Institute of Agrobiological Sciences (Tsukuba, Japan), and other silkworm strains C108, pnd, pS and pe re911 were maintained in the Genetic Resource Center, National Institute of Agrobiological Sciences. Silkworms were reared on mulberry leaves or on an artificial diet (Nihon Nosan Kogyo, Yokohama, Japan) under a 16-h light/8-h dark photoperiod at 25 °C. The wild-type strain of T. castaneum is derived from a strain that was provided from the National Food Research Institute (Tsukuba, Japan) and was raised on whole wheat flour at 30 °C.
Positional cloning of pe
For recombination mapping, six F1 heterozygous males, which were obtained from a single-pair cross between a C108 wild-type female and a pe re911 mutant male, were each backcrossed to a pe re911 mutant female. A total of 656 BC1 eggs (282 normally pigmented eggs, 260 white eggs and 114 red eggs) were used for analyses. Genomic DNA was extracted from parent moths, F1 moths and from each BC1 neonate larva using DNAzol solution (Invitrogen, Carlsbad, CA, USA). For genetic analysis, 14 single-nucleotide polymorphism (SNP) markers previously reported on chromosome 5 (Yamamoto et al., 2008) and 16 new SNP markers that were constructed for this study were used. The primers for each SNP marker are listed in Supplementary Table S1.
Phylogenetic analysis
Deduced amino acid sequences of cardinal gene homologs were aligned using the Muscle program implemented in MEGA6 (Tamura et al., 2013). Molecular phylogenetic analysis was conducted by neighbor-joining and maximum-likelihood methods using MEGA6. The evolutionary distances were computed using the JTT or Poisson model for neighbor-joining or maximum-likelihood analysis, respectively. The confidence of the phylogenetic lineages was assessed by bootstrap analysis of 1000 resamplings.
RNA sequencing (RNAseq) experiment
Total RNA was isolated from at 0, 24, 48 and 72 h after eggs were laid from two wild-type B. mori strains (p50T and C108) and a pe mutant strain pee20 that is different from the pe re911 strain used for linkage analysis. ISOGEN (Nippon Gene, Tokyo, Japan) and RNAeasy mini kit (Qiagen, Dusseldorf, Germany) were used for the RNA extraction from egg samples. RNAseq libraries were prepared using TruSeq Sample Preparation Kit v2 (Illumina Inc., San Diego, CA, USA), and sequenced using 100 bp paired-end strategy on a Hiseq 2500 sequencer (Illumina). Mapping to the most recent B. mori gene list (Suetsugu et al., 2013) was performed with BWA-mem software using MASER pipeline provided by the Cell Innovation Project team at the National Institute of Genetics (http://cell-innovation.nig.ac.jp/public/contents/service_en.html#pf_maser). The relative expression of each gene was estimated to calculate fragments per kilobase of exon per million (FPKM) values. The SNPs of the genes were analyzed by Integrative Genomics Viewer. The RNAseq data set is deposited in DDBJ (DNA Data Bank of Japan) under accession number DRA003068.
RNAi experiments
A small interfering RNA (siRNA) targeted to Bm-cardinal was designed using siDirect program version 2.0 (http://sidirect2.rnai.jp/) (Naito et al., 2009), following the guidelines reported in Yamaguchi et al. (2011). The sequences of the siRNA are as follows: pe-1 sense strand, 5′-GCGAUUACAGCACUUUCAAUG-3′; antisense strand, 5′-UUGAAAGUGCUGUAAUCGCUU-3′. We used the following siRNA for Bm-white as reported in Ando and Fujiwara, 2013; sense strand, 5′-CAUUUAUGGCCCAAAACGUUA-3′; antisense strand 5′-ACGUUUUGGGCCAUAAAUGAA-3′. To produce templates for double stranded RNA (dsRNA) synthesis the following primers were used for T. castaneum cardinal: Tccd-F3 5′-GACGATGTCAGGGGGTGAAC-3′ and Tccd-R2 5′-GTTATAGCCAACTAATCCATGGTC-3′.
For embryonic RNAi of B. mori, 2–3 nl of 100 μM siRNA was injected into eggs 3–5 h after laying as reported previously (Osanai-Futahashi et al., 2012). For larval and pupal RNAi of B. mori, an electroporation-mediated method was performed as reported by Ando and Fujiwara, 2013. The larval epidermis of the third- or fourth-instar pS strain and the compound eye region of day 1–3 pupae were injected with 0.25–0.5 μl of 100 μM siRNA solution. Immediately after injection, platinum electrodes and droplets of gel for Ultrasound GELH-250 (Nepagene, Ichikawa, Japan) were placed nearby and five pulses of 45 V (280 ms s−1) were applied by the electroporator Cure-Gene (CellProduce Co., Ltd, Tokyo, Japan). To perform RNAi using the beetle T. castaneum, double-stranded RNAs were prepared and 0.5 μg μl−1 of dsRNA solution was injected into the abdomens of last- or penultimate-instar larvae as described previously (Osanai-Futahashi et al., 2012).
Knockout experiment using TALEN
TALEN knockout experiments were conducted as reported previously (Takasu et al., 2013). A manual search was performed for a TALEN target site for Bm-cardinal in a conserved region among p50T, C108, and pee20 strains. 5′-GCTGAGGGTCGGGAG-3′ and 5′-TTGGATCAGATGGAAGGA-3′ in exon 8 of Bm-cardinal, which is a part of the peroxidase domain, were selected as the recognition sequences. The TAL portions were prepared by Golden Gate TALEN and TAL Effector Kit (Addgene, Cambridge, MA, USA), following the method described by Cermak et al. (2011), using an in-vitro expression vector pBlue-TAL. The constructed plasmids were purified by HiSpeed Plasmid Midi kit (Qiagen), linearized by XbaI, treated with proteinase K, extracted with phenol/chloroform/isoamyl alcohol (25:24:1) and chloroform, precipitated with ethanol and washed with 70% ethanol three times. TALEN mRNAs were transcribed by mMESSAGE mMACHINE kit (Applied Biosystems, Foster City, CA, USA), followed by lithium chloride precipitation and were then washed with 70% ethanol three times. The resulting TALEN mRNAs were dissolved into injection buffer (0.5 mM phosphate buffer (pH 7.0), 5 mM KCl) to achieve a final concentration of 0.5 mg ml−1 each, and were microinjected into non-diapausing B. mori pnd (wild-type egg color) strain 3–5 h after laying. Of the 192 injected eggs, 105 hatched and 47 became adults. Sixteen G0 males were mated with sibling G0 females to obtain G1 generation eggs, and 10 of the G0 males were re-mated with diapausing pee20 females for complementation test. The eggs for complementation test were subjected to acid treatment 24 h after laying to prevent entering diapause. For genotyping mutations, genomic DNA was isolated from adult heads of moths emerged from pink eggs from the G0 and pee20crosses, and G1 moths, using Maxwell 16 Tissue DNA purification Kit (Promega, Fitchburg, WI, USA) following the manufacturer’s instructions. To identify the indels introduced by TALENs, the targeted region was amplified by PCR with primers 5′-AACCAAGCCACAGCCTTTATC-3′ and 5′-TGATTCAAAACAATAGCGTCCTT-3′, and directly sequenced by ABI prism 3130 Genetic Analyzer (Applied Biosystems).
Quantitative reverse transcriptase-PCR
Total RNA from B. mori eggs was extracted using ISOGEN (Nippon Gene) and used as a template for synthesis of cDNA with a first-strand cDNA synthesis kit (GE Healthcare, Buckinghamshire, UK). The expression level of Bm-cardinal mRNA was quantified using a Light-Cycler 1.5 (Roche, Basel, Switzerland). The B. mori rpL3 gene was analyzed to normalize transcript levels. Quantitative PCR was performed in duplicate. The P-values were obtained using Student’s t-test. Primers used for quantitative PCR are listed in Supplementary Table S2.
Pigment extraction
Ommochrome pigment was directly extracted from four pairs of B. mori compound eyes by 100 μl of 1% hydrochloric acid in methanol (v/v). Because we found that decarboxylated xanthommatin was removed by methanol wash, we did not conduct washes before pigment extraction. After centrifugation, supernatants were collected and used for analyses.
Thin-layer chromatography
TLC was performed by spotting 20 μl of the compound eye extracts on to precoated silica gel plates with concentrating zone (60 F254, Merck, Billerica, MA, USA). They were developed using the upper layer of collidine/lutidine/water (1:1:2, v/v) as solvent (Ajami and Riddiford, 1971; Sawada et al., 2000). The autofluorescence of the separated pigments was observed by 302 nm irradiation using a minitransilluminator (Funakoshi, Tokyo, Japan). 3-hydroxykynurenine was purchased from Sigma (St Louis, MO, USA), and synthesized xanthommatin and decarboxylated xanthommatin were kindly provided by Dr Hiroaki Mano (National Institute for Basic Biology, Okazaki, Japan).
High-pressure liquid chromatography–mass spectrometry
HPLC-MS analysis was conducted following the method used for analysis of Junonia coenia wing ommochrome pigments (Daniels and Reed, 2012). The experiment was conducted by using a Hewlett Packard HP 1100 MSD system fitted with a CAPCELL PAK C18 TYPE UG80 reversed-phase column (3.0 mm ϕ × 150 mm, 5 μm particle size, Shiseido, Tokyo, Japan), running on a linear gradient protocol. The primary solvent was 0.5% acetonitrile with 0.2% acetic acid and increased linearly to 95% acetonitrile in 0.2% acetic acid in 15 min at a flow rate of 0.5 ml min−1. Ultraviolet/visible spectra were monitored with a photodiode array detector. Electrospray ionization in the positive mode was used for the MS detection.
Results
Positional cloning of the pe locus
To identify the candidate region for the pe gene, we conducted a genetic linkage analysis using a high-resolution SNP linkage map (Yamamoto et al., 2008) and silkworm genome sequence information (The International Silkworm Genome Consortium, 2008). First, we obtained F1 heterozygous males from a cross between a single male of pe re double-mutant strain pe re911 and a single female of a wild-type egg and eye colored strain C108. We backcrossed the F1 heterozygous males with pe re911 females, and obtained 282 normally pigmented BC1 eggs (+pe+re/pe re), 114 red BC1 eggs (+pere/pe re) and 260 white BC1 eggs (pe re/pe re or pe +re/pe re). It should be noted that red BC1 eggs are homozygous for the re allele but heterozygous for the pe allele, as pe gene functions above re. By analyzing the SNPs of 656 BC1 eggs, we narrowed down the pe-linked region to within 258 kb on chromosome 5, from position 1 156 521 to 1 414 291 (Supplementary Figure S1 and Supplementary Table S1). Within this region, 17 and 12 genes were predicted by the most recent B. mori gene list (Suetsugu et al., 2013) and canonical gene model (The International Silkworm Genome Consortium, 2008), respectively (Figure 2a).
Positional cloning of the pe locus. (a) A physical map of the pe-linked region. The pe-linked region was narrowed within the 258 kb region between SNP markers pe-080514-L-R7_F1 and pe-080514-R-R18_F20 (Supplementary Table S1). In this region, there are 17 and 12 predicted genes of recently revised gene model (Suetsugu et al., 2013) and canonical gene model (numbers in parenthesis, (The International Silkworm Genome Consortium, 2008)). (b) Expression analysis of 17 predicted genes within the pe-linked region and 7 ommochrome-related genes by RNAseq. The numbers on the x axis indicate hours after oviposition. The numbers on the y axis indicate the level of relative expression (FPKM values). The dark gray, light gray and diagonally lined column indicate the p50T (wild type), C108 (wild type) and pee20 strain, respectively.
Comprehensive transcriptome analysis of wild-type and pe eggs
As the morphological differences between pe and wild-type eggs become distinct 48–72 h after the eggs are laid, the responsible gene is likely to be expressed during or just before this period. We therefore compared the expression pattern and deduced amino acid sequences of all 17 candidate genes among two wild-type strains p50T and C108, and pee20 mutant strain, which is a different pe strain from pe re911, by RNAseq at 0, 24, 48 and 72 h after eggs were laid. In both wild-type and pe strains, seven genes (Gene2414, 17856, 2416, 2417, 17857, 2424 and 2425) and two genes (Gene2415 and 2426) were hardly (FPKM <5) or only weakly expressed (FPKM <20), respectively (Figure 2b). As pe egg color phenotype is not subject to maternal inheritance, we excluded Genes 2418, 17858, 17859, 2420 and 2423 from the candidate genes, because their mRNA detected immediately post laying were very likely to be maternally derived and should cause maternal effect if they were the responsible gene. Expression patterns of two genes (Gene2421 and 2422) were very similar between wild-type and pe strains and no nucleotide changes were found in the pe strain. In the remaining gene (Gene2419), the level of expression was elevated at ∼24 h after laying, just before egg pigmentation occurred, similar with other ommochrome genes such as Bm-re, Bm-scarlet and Bm-white (Figure 2b) (Tatematsu et al., 2011; Osanai-Futahashi et al., 2012). For Bm-white, we summed the FPKMs of two predicted genes that were both alleles of Bm-white derived from different strains. Because maternally synthesized 3-hydroxykynurenine is mainly used for egg pigmentation, the low expression level of vermilion and cinnabar at this period was consistent. We found two paralogous genes for kynurenine formamidase in B. mori (Gene8740 and 8743), and only Gene8743 was expressed around this stage. We found two missense mutations in Gene2419 of the pee20 strain (Figures 3a and d,Supplementary Figure S2). We therefore determined the open reading frame sequence of Gene2419 in another pe mutant strain, pe re911 (hereafter referred to simply as pe911), which was originally used for linkage analysis, and detected one shared missense mutation (arrowhead in Figure 3d, Supplementary Figure S2). This suggested that the missense mutation in Gene 2419 is a strong candidate for the cause of pe phenotype. Although in the RNAseq experiment pee20 had twice the higher expression at 24 h post oviposition, no significant differences were found between wild-type p50T and pee20 or pe911 strains by quantitative reverse transcriptase-PCR (t-test; P=0.25 and 0.62, respectively) (Supplementary Figure S3).
Structure and phylogenetic relationship of the Bm-cardinal gene. (a) Genomic structure of the Bm-cardinal gene. The open reading frame is indicated by solid boxes, the untranslated region is indicated by open boxes and introns are indicated by diagonal lines. The tryptophan to arginine mutation that is specific for the two pe strains is located in exon 9. Exon 13 is mapped in an unmapped scaffold Bm_scaf1910; thus, the exact size of adjoining introns is unknown. (b) The predicted gene structure of the Bm-cardinal gene. The transmembrane domain was predicted by SOSUI (Hirokawa et al., 1998). (c) Phylogenetic relationship of the cardinal gene homolog in insects. Homo sapiens myeloperoxidase, eosinophil peroxidase and peroxidase gene from B. mori and D. plexippus with the second highest e-value by blastp search using Bm-cardinal protein were included as outgroups. A neighbor-joining phylogeny inferred from 1735 aligned amino acid sites is shown, and maximum-likelihood phylogeny was found to exhibit substantially similar topologies. On each node, bootstrap values are indicated in the order of neighbor-joining method/maximum-likelihood method. Bootstrap values over 50% are indicated. Accession numbers or annotation IDs are in brackets. (d) Alignment of insect cardinal homologs. The tryptophan to arginine mutation shared by the two pe strains is indicated by arrowheads, and the aspartic acid to alanine mutation that is specific to pee20 strain is indicated by asterisk.
Gene 2419 encodes a peroxidase gene orthologous to cardinal
Gene2419 encoded a haem peroxidase, the ortholog of the cardinal gene in D. melanogaster (hereafter referred to as Bm-cardinal) (Figure 3c). Notably, the cardinal mutant of D. melanogaster shows delayed eye pigmentation because of slow conversion of 3-hydroxykynurenine to xanthommatin (Harris et al., 2011), thus indicating that Cardinal mediates this process. The predicted cardinal gene product has a transmembrane region and a peroxidase homology domain (Harris et al., 2011), and the two mutations in the Bm-cardinal gene of pee20 were located within the peroxidase domain (Figures 3a and b). Molecular phylogenetic analysis revealed that insect cardinal genes formed monophyletic cluster (Figure 3c) and amino acid sequences of the peroxidase domain were highly conserved among insects (Figure 3d). The tryptophan residue, which was converted to arginine in the two pe strains, was widely conserved in holometabolous insects (arrowhead in Figure 3d), whereas the amino acid at position of the pee20-specific mutation was not conserved (asterisk in Figure 3d). These results support the hypothesis that the shared mutation in the Bm-cardinal gene is a strong candidate for the cause of the pe phenotype.
Functional analysis of Bm-cardinal gene by RNAi
To investigate whether the Bm-cardinal gene is essential for normal egg pigmentation, we performed embryonic RNAi experiments. Most eggs injected with Bm-cardinal siRNA failed to turn into the normal purplish-brown color, but became white like the eggs of the pe mutant (Figure 4a). In contrast, all eggs injected with the injection buffer developed into the normal dark color (Figure 4a). We next analyzed the effect of RNAi on adult compound eyes. Because simple systemic RNAi by injection of dsRNA does not work in the larval and pupal period in Lepidopteran species, including B. mori, in general (Terenius et al., 2011), we conducted recently developed electroporation-mediated RNAi as reported by Ando and Fujiwara (2013). We found that one side of compound eyes became reddish by electroporation of Bm-cardinal siRNA, like the pe mutant, whereas it became white by electroporation of Bm-white siRNA like w-3 (Bm-white) mutant (Figure 4b). All individuals injected with the injection buffer had the normal dark compound eyes (Figure 4b).
Knockdown of the Bm-cardinal gene by RNAi. (a) The effect of RNAi on egg pigmentation. Bm-cardinal siRNA or injection buffer was injected into eggs at 3–6 h post oviposition, and eggs were then incubated at 25 °C. The injected eggs were photographed after 3 days. (b) The effect of RNAi on the pigmentation of adult compound eyes. One side of the eyes (arrowheads) was subjected to electroporation-mediated RNAi at day 0 pupa. (c) The effect of RNAi on pigmentation around eyespot markings of the fifth-instar larva. One side of eyespot markings (arrowheads) was subjected to electroporation-mediated RNAi at the fourth or third instar.
Ommochromes are also involved in red and brown pigmentation of Lepidopteran larval markings. In the pS strain, conspicuous red markings are recognized around eyespot markings of final-instar larva. We found that this red marking disappears with individuals on a pe background (Figure 1). We therefore examined the effect of Bm-cardinal RNAi on larval pigmentation. By using electroporation-mediated RNAi, we confirmed that red pigmentation could be suppressed by introduction of either Bm-cardinal siRNA or Bm-white siRNA, but not with injection buffer only (Figure 4c), suggesting that these genes are essential for the formation of red markings on larval epidermis. Overall, the pe phenotype was reproduced in coloration of eggs, adult eyes and larval epidermis by Bm-cardinal RNAi.
Functional analysis of Bm-cardinal gene using TALEN
We next performed genetic knockout experiments for Bm-cardinal by TALEN. TALEN target sites were designed in exon 8 (Figure 5a), 132 nucleotides upstream of the pe-specific mutation (Figure 2b). A large portion of the eggs injected with TALEN mRNA showed a mosaic color phenotype between white and normal purple brown to varying degrees (Figures 5b and c). In the G0 generation, we observed two adults with red eyes like pe, 20 adults with mosaic colored eyes and 25 adults with black eyes (Figures 5b and c). We obtained 16 G1 egg batches from G0 sibling pairing, among which 15 batches contained pink eggs (Supplementary Figure S4). Notably, pink egg coloration is because of prolonged preservation of non-diapausing eggs at low temperature, and was not observed in non-diapausing eggs left to develop at room temperature, as well as in diapaused eggs. All adults derived from pink G1 eggs had pink-red eyes like the pe mutant (Figure 5d). We also found that neonatal larvae from pink G1 eggs, as well as pe mutants, have lighter coloration in their abdominal prolegs (Figure 5e), suggesting that the brownish color around the prolegs in the wild type are caused by ommochrome pigments.
Disruption of the Bm-cardinal gene using designed TALENs. (a) The genomic structure of the Bm-cardinal gene and the TALEN binding sites. The TALEN target sites are located in exon 8. (b) The mosaic coloration phenotype of G0 eggs and adult compound eyes injected with TALEN mRNA targeting Bm-cardinal. TALEN mRNA was injected into eggs 3–6 h post oviposition, and eggs were then incubated at 25 °C. The eggs and compound eyes showed various ranges of mosaic coloration. Injected eggs were photographed after 3 days. (c) Summary of the G0 phenotype resulting from TALEN injection. (d) Adult compound eyes of the G1 generation. Eggs obtained from sibling mating of G0 moths derived from eggs injected with TALEN mRNA targeting Bm-cardinal, and an adult derived from a pink G1 egg is shown. (e) Wild-type neonatal larva (left), G1 generation of Bm-cardinal targeting TALEN experiment, derived from a pink egg (middle), and a pee20mutant strain. Both pe- and Bm-cardinal-disrupted neonatal larva lack brown pigmentation in the abdominal legs. (f) Compound eyes of an adult derived from pink egg produced by crossing pee20 females with G0 male moths targeted for Bm-cardinal. The specimen in this picture corresponds to CT-E-F-1 of (g). (g) Sequences of small, induced insertions and deletions in Bm-cardinal for 13 G1 mutants, and 3 adults from the complementation test. The inserted sequences are underlined. Only the alleles with TALEN-mediated indels are shown for the complementation test (the pee20-derived allele has the same sequence as wild type (WT) in this region).
To confirm TALEN-mediated gene disruption, we extracted genomic DNA from 16 G1 adults with pink eyes from batches 1 and 2 (Supplementary Figure S3), and sequenced the 200 bp region containing the TALEN target sites. Direct sequencing of the PCR product showed that three individuals had overlapping sequences just after the TALEN target sites, possibly heterozygous for indels. All of the remaining 13 individuals had non-overlapping sequences with a small deletion and/or an insertion (Figure 5g), indicating that the Bm-cardinal gene is disrupted in these individuals.
Complementation test of pe mutant female with Bm-cardinal-deficient male
Results from the expression analyses, RNAi and TALEN experiments strongly suggest that the Bm-cardinal gene corresponds to the pe locus. To confirm this, we performed a complementation test by crossing pe mutant females with Bm-cardinal-deficient G0 males. We obtained 10 batches of eggs, all of which contained pink eggs (Supplementary Figure S5). We found that all adults derived from pink eggs had red eyes like pe (Figure 5f), indicating that the pe phenotype could not be rescued by a Bm-cardinal-deficient male. Furthermore, we extracted genomic DNA from the adult heads and amplified the 200 bp region containing the TALEN target sites; we found that these individuals were heterozygotes of a pee20-derived sequence that is the same as that of the wild-type individuals for this exon, and a sequence with insertions or deletions in the region between the two TALEN target sites (Figure 5g). The above results demonstrate that the Bm-cardinal gene is responsible for the pe phenotype.
Pigment analyses of B. mori adult eyes
To analyze the changes of eye pigment composition between wild type and pe, we extracted ommochrome pigments from adult compound eyes with acidic methanol for pigment analysis (Supplementary Figure S6a). We omitted washes before the pigment extraction to detect decarboxylated xanthommatin (Futahashi et al., 2012). The extracted pigments from pe or Bm-cardinal knockout individuals showed lighter coloration than that from wild-type p50T strain (Supplementary Figure S6a).
In TLC analysis, we analyzed the extracted pigments from p50T and pe eyes, along with 3-hydroxykynurenine, synthesized xanthommatin and synthesized decarboxylated xanthommatin in collidine/lutidine/water (1:1:2) solvent system on silica gel plate. A purple-red band of RF=0.46 was observed in p50T eye extract that was faintly found in pe eye extract and was absent in the standard pigments (Supplementary Figure S6b). This band was in good correspondence with the one of the ommin spots (RF=0.46) reported from saturniid moth eye (Ajami and Riddiford, 1971). We observed autofluorescence in pe eye extract at the similar mobility with 3-hydroxykynurenine (Supplementary Figure S6b), suggesting the accumulation of 3-hydroxykynurenine in the pe eye as reported previously (Inagami, 1954).
We also performed HPLC-MS analyses for acidic methanol extract of eyes from Bm-cardinal knockout individuals and p50T. In comparison with chromatogram and the mass data of the synthesized xanthommatin and decarboxylated xanthommatin, we found that both xanthommatin and decarboxylated xanthommatin were obviously present in p50T eye extracts (Supplementary Figures S7–S10). Although xanthommatin was also detected in Bm-cardinal knockout individuals, it was not a major component based on electrospray ionization mass spectra of the mass peak of corresponding retention time and the extracted ion chromatogram (Supplementary Figures S11 and S12). Decarboxylated xanthommatin was hardly detected in Bm-cardinal knockout eye extract (Supplementary Figures S11 and S12). Notably, several uncharacterized substances seem to be major components of Bm-cardinal knockout extract around the xanthommatin and decarboxylated xanthommatin retention time (Supplementary Figures S11 and S12). These results suggest that both ommin and ommatin composition of Bm-cardinal deficient insect differs largely from that of wild type.
Knockdown of the cardinal ortholog gene in the beetle T. castaneum
Like B. mori, most insects have blackish eyes because of ommin pigments (Linzen, 1974; Osanai-Futahashi et al., 2012). To investigate the function of the cardinal gene ortholog in other insects, we performed RNAi of cardinal ortholog in T. castaneum (Tc-cardinal). All pupae treated with Tc-cardinal dsRNA (n=16) did not develop eye pigmentation throughout the pupal stage and newly emerged adults had white compound eyes (Figure 6), similar to individuals in which ommochrome-related genes Tc-vermilion, Tc-white and Tc-scarlet were knocked down (Broehan et al., 2013). All pupae treated with EGFP dsRNA (n=17) had normal eye pigmentation (Figure 6). Notably, the adult compound eyes of individuals treated with Tc-cardinal dsRNA became reddish several days after eclosion (Figures 6b–d), similar to the cardinal mutant of D. melanogaster. These results indicate that the Tc-cardinal gene functions in eye pigmentation and ommochrome biosynthesis in T. castaneum as it does in B. mori.
Knockdown of a cardinal homolog by RNAi in T. castaneum. The last- or penultimate-instar larvae were injected with 170 nl of dsRNA (500 ng μl−1) for the cardinal homolog in T. castaneum (Tccd) or EGFP. (a) Ventral view of pupae at day 4. Arrowheads indicate compound eyes. (b) Ventral view of adult at day 0, or 5 days after eclosion. The specimen of day 0 Tc-cardinal RNAi (left) was photographed just after eclosion. (c) Magnified view of adult compound eyes of (b). (d) Summary of the pattern of eye pigmentation during developmental stages of the RNAi experiments.
Discussion
In this study, we found that a missense mutation in the Bm-cardinal gene is responsible for the silkworm pe locus. The original Drosophila cardinal gene product has been regarded as a phenoxazinone synthase for converting 3-hydroxykynurenine into xanthommatin (Howells et al., 1977; Harris et al., 2011). Although both the Bombyx pe mutant and the Drosophila cardinal mutant seem to accumulate the ommochrome precursor 3-hydroxykynurenine (Supplementary Figure S6a, Inagami, 1954; Howells et al., 1977), it should be noted that they have different eye color phenotypes. Drosophila cardinal has reduced ommochrome levels at eclosion and becomes close to the wild-type compound eye color during aging, but the pink eyes of the Bombyx pe mutant do not become black like those of the wild type. Notably, eye pigments of most insects, including Bombyx, have been reported to be a mixture of purple ommin pigments and reddish xanthommatin, which gives eyes black coloration in many insects, whereas ommin is absent in eyes of higher Dipteran species including Drosophila (Linzen, 1974). In the present study, we found that decarboxylated xanthommatin is also a major pigment of wild-type B. mori eyes (Supplementary Figure S10). Because decarboxylated xanthommatin can be removed by methanol wash, it has been missed in the previous study. By TLC and HPLC-MS results, we confirmed obvious decrease of not only red-purple pigment that is a good candidate for ommin, but also xanthommatin and decarboxylated xanthommatin in Bm-cardinal-deficient insect eyes. Instead, we found that substances with different molecular weights are the majority of the mass peak of the same retention time with xanthommatin and decarboxylated xanthommatin, suggesting that other substances, possibly pigments, are produced in these insects. We could not identify these substances as well as ommins by HPLC-MS, and this deserves future studies.
In addition to eye and egg coloration, we also found that Bm-cardinal is essential for ommochrome pigmentation in the larval epidermis. A recent study on Bombyx ommochrome and melanin larval mutant quail hypothesized that BGIBMGA006740 (Gene5459 in Suetsugu et al., 2013) is the phenoxazinone synthetase gene (Nie et al., 2014). Because Gene5459 was hardly expressed around the egg pigmentation period (Figure 2b), and has sequence similarity with the laccase2 gene, it may function in melanin pigmentation rather than ommochrome pigmentation. The observed phenotype of the pe mutant and the results from RNAi experiments suggests that Bm-cardinal is involved in ommochrome biosynthesis in larval epidermis, at least for the red markings in the pS strain (Figures 1 and 4c). Although ommochrome pigments are absent in white Bombyx wings, they are present in wings of Nympalidae butterflies such as Heliconius species and Precis coenia (Butenandt and Schafer, 1962; Gilbert et al., 1988; Nijhout, 1997), and phenoxazinone synthetase activity has been detected in pupal P. coenia wings (Nijhout, 1997). We confirmed the presence of cardinal gene homolog in the developing wing expressed sequence tag data set of Heliconius erato (IC33431AfEcon6544) (Hines et al., 2012) by tBlastn search (http://butterflygenome.org/), and the cardinal gene homolog was also reported in the transcriptomes of Hawaiian happy-face spiders (Croucher et al., 2013). It may be of interest to determine whether the cardinal gene ortholog is involved in coloration of butterfly wings and spider bodies.
Notably, the effects of Bm-cardinal suppression on pigmentation varied among tissues. As mentioned above, red ommochrome coloration was always observed in compound eyes of pe mutants and the individuals that had Bm-cardinal gene experimentally knocked out/down with TALEN and RNAi, respectively, whereas embryonic RNAi produced white eggs in general and both pe mutants and individuals treated with Bm-cardinal siRNA suppressed red pigmentation of larval epidermis. This suggests that the absence of Bm-cardinal activity results in complete or strong repression of conversion of 3-hydroxykynurenine into ommochrome pigments in eggs and larval epidermis, whereas it results in altered ommochrome synthesis in adult compound eyes. Difference of the coloration phenotype between tissues in cardinal mutants is also reported in D. melanogaster. Compound eyes of cardinal mutants produce xanthommatin during aging, but their ocelli are colorless. One explanation to account for variation of the pigmentation phenotype between tissues is the difference in the efficiency of 3-hydroxykynurenine auto-oxidation into xanthommatin that is known to occur spontaneously in vitro (Wiley and Forrest, 1981). Considering the coloration phenotype and the pigment analysis results, we propose that the loss of cardinal activity basically represses the ommochrome synthesis and possibly substances with reddish coloration similar with xanthommatin are produced by auto-oxidization in the mutant eyes in a delayed manner. Notably, when the non-diapausing destined eggs were preserved for several weeks at low temperature, pe and Bm-cardinal TALEN knockout eggs turned white to pink (Supplementary Figure S4), and this was not observed in diapaused eggs. One contributing factor that prevents cardinal independent red pigment formation may be the recently reported Bombyx homolog of human oxidation resistance 1 (OXR1) protein that has higher mRNA expression in diapause eggs than in non-diapausing eggs (Kobayashi et al., 2014). From biochemical experiments with Drosophila wild-type and eye color mutants, it has been proposed that xanthommatin itself also acts as an oxidizing agent for its own precursor 3-hydroxykynurenine, and is concomitantly converted into its reduced form, dihydroxanthommatin, which is the major form in insect tissues (Wiley and Forrest, 1981). As the catalytic activity by xanthommatin is absent at the onset of xanthommatin synthesis, it is most likely that peroxidase, which is encoded by the cardinal gene, triggers xanthommatin synthesis in the pupal ommatidia of both in B. mori and D. melanogaster (Harris et al., 2011).
In addition to B. mori, we found that the cardinal ortholog in T. castaneum was also involved in normal eye pigmentation (Figure 6). Notably, the newly emerged Tc-cardinal RNAi beetle displayed white eyes, suggesting that xanthommatin synthesis occurred after eclosion as it occurred in the cardinal mutant of D. melanogaster (Figure 6). The differences in pigmentation timing in the cardinal mutant among species coincides with their pupal periods; D. melanogaster has a short pupal period of 4 days and pigment synthesis starts after 49 h, T. castaneum has a short pupal period of 6 days and the ommatidia starts to color at day 2 and B. mori has a pupal period of 12–14 days and compound eye coloration can be detected at 5 days before eclosion. Because of a short pupal period, cardinal-independent red pigment synthesis may be incomplete at the time of eclosion in the cardinal-deficient Drosophila and Tribolium individuals.
The conservation of cardinal homolog function in Tribolium, Bombyx and Drosophila, and the widespread distribution of cardinal homologues in insects (Figure 3c), suggest that cardinal functions in black eye coloration in many insect species. Notably, some mutants are quite similar to pe mutants. In T. castaneum, the cardinal homolog is located in the same linkage group X that harbors several red- or pink-eyed mutants such as red-l, peach, pink Ndg and pink Tiw (Lorenzen et al., 2002). The Culex pipiens mutant crimson is also a recessive mutant with colorless ocelli at first-instar larva that gradually turn red during larval development (Rasgon and Scott, 2004). Eye pigments detected by the head squash method of teneral crimson adults are yellow and turn to red-brown with age, whereas teneral and aged wild-type eye pigments are black, and the teneral adult eyes have excess 3-hydroxykynurenine. Both the tangerine mutant of blowfly Lucilia cuprina and the chartreuse mutant of honeybee Apis mellifera have excess 3-hydroxykynurenine and impaired ommochrome pigment synthesis (Dustmann, 1968; Linzen, 1974; Summers and Howells, 1980). Comparative studies on these mutants should increase our understanding of ommochrome biosynthesis.
Data archiving
Data available from the DDBJ repository: accession number DRA003068.
Accession codes
References
Ajami AM, Riddiford LM . (1971). Identification of an ommochrome in the eyes and nervous systems of saturniid moths. Biochemistry 10: 1451–1455.
Ando T, Fujiwara H . (2013). Electroporation-mediated somatic transgenesis for rapid functional analysis in insects. Development 140: 454–458.
Ariga I, Murakami A . (1975). Induction of recombinants by mitomycin C in oogenesis of the silkworm (Bombyx mori L.). J Seric Sci Jpn 44: 154–160.
Ariga I, Murakami A . (1976). Recombinogenic effects of ethyl methanesulfonate on the silkworm oocytes. J Seric Sci Jpn 45: 219–224.
Banno R, Shimada T, Kajiura Z, Sezutsu H . (2010). The silkworm-an attractive BioResource supplied by Japan. Exp Anim 59: 139–146.
Broehan G, Kroeger T, Lorenzen M, Merzendorfer H . (2013). Functional analysis of the ATP-binding cassette (ABC) transporter gene family of Tribolium castaneum. BMC Genomics 14: 6.
Butenandt A, Biekert E, Linzen B . (1958). [Ommochromes. XIV. Distribution of ommins in animal kingdom]. Hoppe Seylers Z Physiol Chem 313: 251–258.
Butenandt A, Schafer W . (1962) Ommochromes. In: Gore TS (ed) Recent Progress in the Chemistry of Natural and Synthetic Coloring Matters and Related Fields. Academic Press: New York, NY, USA, pp 13–33.
Cermak T, Doyle EL, Christian M, Wang L, Zhang Y, Schmidt C et al. (2011). Efficient design and assembly of custom TALEN and other TAL effector-based constructs for DNA targeting. Nucleic Acids Res 39: e82.
Croucher PJ, Brewer MS, Winchell CJ, Oxford GS, Gillespie RG . (2013). De novo characterization of the gene-rich transcriptomes of two color-polymorphic spiders, Theridion grallator and T. californicum (Araneae: Theridiidae), with special reference to pigment genes. BMC Genomics 14: 862.
Daniels EV, Reed RD . (2012). Xanthurenic acid is a pigment in Junonia coenia butterfly wings. Biochem Syst Ecol 44: 161–163.
Dustmann JH . (1968). Pigment studies on several eye-colour mutants of the honey bee, Apis mellifera. Nature 219: 950–952.
Futahashi R, Kurita R, Mano H, Fukatsu T . (2012). Redox alters yellow dragonflies into red. Proc Natl Acad Sci USA 109: 12626–12631.
Gilbert LE, Forrest HS, Schultz TD, Harvey DJ . (1988). Correlations of ultrastructure and pigmentation suggest how genes control development of wing scale of Heliconius butterflies. J Res Lepid 26: 141–160.
Harris DA, Kim K, Nakahara K, Vasquez-Doorman C, Carthew RW . (2011). Cargo sorting to lysosome-related organelles regulates siRNA-mediated gene silencing. J Cell Biol 194: 77–87.
Hines HM, Papa R, Ruiz M, Papanicolaou A, Wang C, Nijhout HF et al. (2012). Transcriptome analysis reveals novel patterning and pigmentation genes underlying Heliconius butterfly wing pattern variation. BMC Genomics 13: 288.
Hirokawa M . (1993). Induction of males from meiotic parthenogenetic eggs and haploid lethality of these eggs in the silkworm, Bombyx mori. J Seric Sci Jpn 62: 105–110.
Hirokawa M . (1996). Detection of mixoploid by using egg color genes in parthenotes of the silkworm, Bombyx mori. J Seric Sci Jpn 65: 390–391.
Hirokawa T, Boon-Chieng S, Mitaku S . (1998). SOSUI: classification and secondary structure prediction system for membrane proteins. Bioinformatics 14: 378–379.
Howells AJ, Summers KM, Ryall RL . (1977). Developmental patterns of 3-hydroxykynurenine accumulation in white and various other eye color mutants of Drosophila melanogaster. Biochem Genet 15: 1049–1059.
Inagami K . (1954). (VII) On the metabolism of 3-hydoxykynurenine in the egg. J Seric Sci Jpn 24: 91–96.
Kawase S, Aruga H . (1954). Biochemical genetics on the pigment in eggs and compound eyes in the silkworm. Jpn J Genet 29: 114–121.
Kobayashi N, Takahashi M, Kihara S, Niimi T, Yamashita O, Yaginuma T . (2014). Cloning of cDNA encoding a Bombyx mori homolog of human oxidation resistance 1 (OXR1) protein from diapause eggs, and analyses of its expression and function. J Insect Physiol 68: 58–68.
Koga N, Osanai M . (1967). Der Gehalt an Tryptophan, Kynurenin, 3-Hydroxy-Kynurenin und Ommochrome bei den überwinternden Eiern des Seidenspinners Bombyx mori während der Entwicklung. Hoppe Seylers Z Physiol Chem 348: 979–982.
Komoto N, Quan GX, Sezutsu H, Tamura T . (2009). A single-base deletion in an ABC transporter gene causes white eyes, white eggs, and translucent larval skin in the silkworm w-3(oe) mutant. Insect Biochem Mol Biol 39: 152–156.
Linzen B . (1974). The tryptophan-ommochrome pathway in insects. Adv Insect Physiol 10: 117–246.
Lorenzen MD, Brown SJ, Denell RE, Beeman RW . (2002). Cloning and characterization of the Tribolium castaneum eye-color genes encoding Tryptophan Oxygenase and Kynurenine 3-Monooxygenase. Genetics 160: 225–234.
Naito Y, Yoshimura J, Morishita S, Ui-Tei K . (2009). siDirect 2.0: updated software for designing functional siRNA with reduced seed-dependent off-target effect. BMC Bioinformatics 10: 392.
Nie H, Liu C, Cheng T, Li Q, Wu Y, Zhou M et al. (2014). Transcriptome analysis of integument differentially expressed genes in the pigment mutant (quail during molting of silkworm, Bombyx mori. PLoS One 9: e94185.
Nijhout HF . (1997). Ommochrome pigmentation of the linea and rosa seasonal forms of Precis coenia (Lepidoptera: Nymphalidae). Arch Insect Biochem 36: 215–222.
Osanai-Futahashi M, Tatematsu K, Yamamoto K, Narukawa J, Uchino K, Kayukawa T et al. (2012). Identification of the Bombyx red egg gene reveals involvement of a novel transporter family gene in late steps of the insect ommochrome biosynthesis pathway. J Biol Chem 287: 17706–17714.
Pepling M, Mount SM . (1990). Sequence of a cDNA from the Drosophila melanogaster white gene. Nucleic Acids Res 18: 1633.
Quan GX, Kobayashi I, Kojima K, Uchino K, Kanda T, Sezutsu H et al. (2007). Rescue of white egg 1 mutant by introduction of the wild-type Bombyx kynurenine 3-monooxygenase gene. Insect Sci 14: 85–92.
Rasgon JL, Scott TW . (2004). Crimson: a novel sex-linked eye color mutant of Culex pipiens L. (Diptera: Culicidae). J Med Entomol 41: 385–391.
Sawada H, Nakagoshi M, Mase K, Yamamoto T . (2000). Occurrence of ommochrome-containing pigment granules in the central nervous system of the silkworm, Bombyx mori. Comp Biochem Physiol B Biochem Mol Biol 125: 421–428.
Searles LL, Ruth RS, Pret AM, Fridell RA, Ali AJ . (1990). Structure and transcription of the Drosophila melanogaster vermilion gene and several mutant alleles. Mol Cell Biol 10: 1423–1431.
Searles LL, Voelker RA . (1986). Molecular characterization of the Drosophila vermilion locus and its suppressible alleles. Proc Natl Acad Sci USA 83: 404–408.
Suetsugu Y, Futahashi R, Kanamori H, Kadono-Okuda K, Sasanuma S, Narukawa J et al. (2013). Large scale full-length cDNA sequencing reveals a unique genomic landscape in a lepidopteran model insect, Bombyx mori. G3 (Bethesda) 3: 1481–1492.
Summers KM, Howells AJ . (1980). Functions of the white and topaz loci of Lucilia cuprina in the production of the eye pigment xanthommatin. Biochem Genet 18: 643–653.
Takasu Y, Sajwan S, Daimon T, Osanai-Futahashi M, Uchino K, Sezutsu H et al. (2013). Efficient TALEN construction for Bombyx mori gene targeting. PLoS One 8: e73458.
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S . (2013). MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol 30: 2725–2729.
Tatematsu K, Yamamoto K, Uchino K, Narukawa J, Iizuka T, Banno Y et al. (2011). Positional cloning of silkworm white egg 2 (w-2) locus shows functional conservation and diversification of ABC transporters for pigmentation in insects. Genes Cells 16: 331–342.
Tearle RG, Belote JM, McKeown M, Baker BS, Howells AJ . (1989). Cloning and characterization of the scarlet gene of Drosophila melanogaster. Genetics 122: 595–606.
Terenius O, Papanicolaou A, Garbutt JS, Eleftherianos I, Huvenne H, Kanginakudru S et al. (2011). RNA interference in Lepidoptera: an overview of successful and unsuccessful studies and implications for experimental design. J Insect Physiol 57: 231–245.
The International Silkworm Genome Consortium. (2008). The genome of a lepidopteran model insect, the silkworm Bombyx mori. Insect Biochem Mol Biol 38: 1036–1045.
Uda H . (1932). On egg coloration of domestic silkworm. Scientific papers dedicated to the tenth anniversary of the Miye College of Agriculture and Forestry 21–42.
Warren WD, Palmer S, Howells AJ . (1996). Molecular characterization of the cinnabar region of Drosophila melanogaster: identification of the cinnabar transcription unit. Genetica 98: 249–262.
Wiley K, Forrest HS . (1981). Terminal synthesis of xanthommatin in Drosophila melanogaster. IV. Enzymatic and nonenzymatic catalysis. Biochem Genet 19: 1211–1221.
Yamaguchi J, Mizoguchi T, Fujiwara H . (2011). siRNAs induce efficient RNAi response in Bombyx mori embryos. PLoS One 6: e25469.
Yamamoto K, Nohata J, Kadono-Okuda K, Narukawa J, Sasanuma M, Sasanuma S et al. (2008). A BAC-based integrated linkage map of the silkworm Bombyx mori. Genome Biol 9: R21.
Acknowledgements
We are grateful to Yutaka Banno (National BioResource Project (NBRP), Kyushu University) for providing pe e20 strain, Kazutoshi Yoshitake for valuable advice on RNAseq data analysis, Toshiya Ando for advice on electroporation-mediated RNAi, Hiroaki Mano for providing synthesized xanthommatin and decarboxylated xanthommatin, Hiroshi Sawada for advice on pigment analysis, Takahiro Shiotsuki for help with TLC analysis, Eiji Okada for providing the pS strain, Kaoru Nakamura and Toshihiko Misawa for rearing silkworms and for technical support on silkworm embryonic injection, Tetsuya Iizuka for providing the pnd strain, rearing silkworms and for valuable advice, Sayaka Kobayashi for assistance with quantitative PCR analyses, Kotaro Takano for helpful comments and anonymous reviewers for comments for improving the manuscript. This work was supported by JSPS KAKENHI Grant Number 26850220 to MO-F, Cooperative Research Grant of the Genome Research for BioResource, NODAI Genome Research Center, Tokyo University of Agriculture and the National BioResource Project, Japan. MO-F is a recipient of a JSPS Research Fellowship for Young Scientists.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on Heredity website
Rights and permissions
About this article
Cite this article
Osanai-Futahashi, M., Tatematsu, Ki., Futahashi, R. et al. Positional cloning of a Bombyx pink-eyed white egg locus reveals the major role of cardinal in ommochrome synthesis. Heredity 116, 135–145 (2016). https://doi.org/10.1038/hdy.2015.74
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/hdy.2015.74
This article is cited by
-
Vermilion and cinnabar are involved in ommochrome pigment biosynthesis in eyes but not wings of Bicyclus anynana butterflies
Scientific Reports (2023)
-
Disruption of kynurenine pathway reveals physiological importance of tryptophan catabolism in Henosepilachna vigintioctopunctata
Amino Acids (2021)
-
Ommochrome pathway genes kynurenine 3-hydroxylase and cardinal participate in eye pigmentation in Plutella xylostella
BMC Molecular and Cell Biology (2020)
-
Comparative analysis of integument transcriptomes identifies genes that participate in marking pattern formation in three allelic mutants of silkworm, Bombyx mori
Functional & Integrative Genomics (2020)
-
Electroporation-mediated RNA interference reveals a role of the multicopper oxidase 2 gene in dragonfly cuticular pigmentation
Applied Entomology and Zoology (2017)