Abstract
The influence of host and parasite genetic background on infection outcome is a topic of great interest because of its pertinence to theoretical issues in evolutionary biology. In the present study, we use a classical genetics approach to examine the mode of inheritance of infection outcome in the crustacean Daphnia magna when exposed to the bacterial parasite Pasteuria ramosa. In contrast to previous studies in this system, we use a clone of P. ramosa, not field isolates, which allows for a more definitive interpretation of results. We test parental, F1, F2, backcross and selfed parental clones (total 284 genotypes) for susceptibility against a clone of P. ramosa using two different methods, infection trials and the recently developed attachment test. We find that D. magna clones reliably exhibit either complete resistance or complete susceptibility to P. ramosa clone C1 and that resistance is dominant, and inherited in a pattern consistent with Mendelian segregation of a single-locus with two alleles. The finding of a single host locus controlling susceptibility to P. ramosa suggests that the previously observed genotype–genotype interactions in this system have a simple genetic basis. This has important implications for the outcome of host–parasite co-evolution. Our results add to the growing body of evidence that resistance to parasites in invertebrates is mostly coded by one or few loci with dominance.
Similar content being viewed by others
Introduction
The notion that both host and parasite genotypes are key determinants of infection outcome underlies much of the evolutionary theory pertaining to host–parasite interactions. Several models used to analyse the influences on and effect of parasitism explicitly rely on this premise. For example, the Red Queen Hypothesis suggests that host–parasite genotype–genotype interactions with a simple genetic basis are important for the maintenance of genetic variation and genetic recombination in the host (Hamilton, 1980; Little and Ebert, 2000). Furthermore, host–parasite genotypic interactions have been implicated in other phenomena, such as the evolution of virulence (Nowak and May, 1994; Grigg and Suzuki, 2003), and may be a significant complicating factor in dealing with infectious diseases in humans (Read and Taylor, 2001; Lambrechts et al., 2005).
Substantial data and clear genetic models already exist on host–parasite genotypic interactions in plants (Thompson and Burdon, 1992 and references therein) and significant inroads have been made into unravelling host–parasite genotypic interactions in invertebrates in a number of systems, including aphid (Acyrthosiphon pisum) and parasitic wasp (Aphidus ervi) (Henter and Via, 1995), snail (Bulinus globosus) and schistosome (Morand et al., 1996; Webster and Woolhouse, 1998), snail (Potamopygrus antipodarum) and trematode (Microphallus species) (Lively and Dybdahl, 2000), bumble bee (Bombus terrestris) and trypanosome (Crithidia bombi) (Schmid-Hempel et al., 1999; Schmid-Hempel and Funk, 2004), Caenorhabditis elegans and soil bacteria (Schulenburg and Ewbank, 2004), and mosquito (Anopheles gambiae) and malarial parasite (Plasmodium falciparum) (Lambrechts et al., 2005). Information on the inheritance of the genotype–genotype interactions is available for very few invertebrate systems. For example, seven main effect quantitative trait locus were reported to affect infection intensity of C. bombi in B. terrestris (Wilfert et al., 2007) and the heritability of strain-specific resistance has been demonstrated in the B. globosus/schistosome system (Webster and Woolhouse, 1998). However, the complexity of the parasite life cycle in many invertebrate systems, which may involve multiple hosts, coupled with the difficulty of isolating either host or parasite clones has put significant hurdles in the way of discovering patterns of inheritance of resistance and susceptibility among different combinations of host and parasite genotypes.
A recent study on the water flea Daphnia magna infected with the castrating bacterial pathogen Pasteuria ramosa described extreme genotype–genotype interactions for infectivity (Luijckx et al., 2011). With evidence for genotypic interactions, fast acting selection (Little and Ebert, 2000) and frequency-dependent selection in natural populations (Decaestecker et al., 2007) this host–parasite system has become one of the prime models for antagonistic co-evolution. However, genetics underlying the genotype–genotype interactions are unknown. In the present study, we use a classical genetic approach to examine the inheritance of resistance in the D. magna–P. ramosa system.
This research differs from previous work on host–parasite interactions with D. magna in several respects (for example, Little and Ebert, 2000; Carius et al., 2001; Little et al., 2006). First, we employ a clone of P. ramosa (single genotype), not field isolates. Field isolates may contain more than one parasite clone (Jensen et al., 2006; Mouton et al., 2007; Ben-Ami et al., 2008; Luijckx et al., 2011). The use of P. ramosa clones negates the complicating factors intrinsic to mixed infections and allows for a more definitive interpretation of experimental results (Luijckx et al., 2011). Second, we use a recently developed attachment test to assess host clones for susceptibility, as assed by attachment of the parasite to the host oesophagus (part of the gut wall). This allows us to separate the step where the parasite attaches to the host, which is believed to be the key step in P. ramosa–D. magna co-evolution, from the other steps in the infection process (encounter and proliferation within the host). In addition, it allows for higher sample sizes than classical infection trials (Duneau et al., 2011). Third, we employ a structured Mendelian approach in which D. magna inbred parental clones, a F1 clone, an array of F2 clones, an array of backcrossed clones and selfed parental clones are used to resolve the genetic pattern of inheritance underlying susceptibility. A further understanding of the genetics of this model system will greatly enhance its use in explaining the factors involved in and the evolutionary implications of host–parasite genotypic interactions.
Materials and methods
Study system
Daphnia magna Straus is a cyclical parthenogenetic, freshwater cladoceran, found in rock pools, small ponds and medium sized lakes. P. ramosa is a bacterial endoparasite of D. magna (Ebert et al., 1996). Transmission occurs when hosts ingest waterborne spores, which attach to the oesophagus, penetrate and subsequently cause infection (Duneau et al., 2011). During the infection millions of spores fill the host body cavity; upon death the spores are released from the decaying cadaver and the cycle begins anew (Ebert et al., 1996). Infection also results in castration, which often occurs before the production of any offspring and therefore entails severe fitness consequences for the host (Ebert et al., 2004).
Host material
Two D. magna individuals were collected from separate rock pools near the Tvärminne Zoological Station in Southern Finland. These rock pools are part of a large D. magna metapopulation. The females were cloned (iso-female lines) under standard conditions, (20±0.5 °C, 16:8 h light:dark cycle and fed with chemostat-cultured algae Scenedesmus obliquus), and then were each selfed (sexual reproduction between clonal male and female offspring) over three generations to create the parental clones used in our study: Fainb3 and Xinb3. The parental clones were crossed and one F1 clone was selfed to create 71 F2 clones. In addition, the F1 clone was backcrossed to parental Fainb3 to create 164 backcrossed clones. Finally, both parent clones (Xinb3 and Fainb3) were selfed. We obtained 22 and 24 offspring clones for selfed Fainb3 and selfed Xinb3, respectively.
The cross to obtain F1 was performed by placing multiple individuals from both parent clones together in 400-ml beakers filled with artificial medium (ADaM, Ebert et al., 1998). Beakers were filled to 90% of their maximum capacity unless otherwise stated. Ephippia containing the sexually produced eggs were removed as they appeared, stored in moist, dark conditions at 4 °C for up to 6 months and then dried on filter paper for up to 3 weeks. Ephippia were then submerged in bleach (5% aqueous solution, household strength) for about 5 min to facilitate the hatching process, washed and placed in 400-ml beakers with medium under strong fluorescent artificial day light. After hatching of sexual eggs, microsatellite markers were used to distinguish hybrid clones from selfed clones (Colson et al., 2009). One hybrid was randomly picked to become the F1 and subsequently selfed to create the F2 clones.
A similar protocol was used to self the F1 and the two parent clones, except that genotyping in these crosses was not necessary. For the backcross, we used a slightly altered approach, as many microsatellites would have been needed to reliably distinguish selfed from outcrossed Daphnia. Fourteen-day-old virgin females of the F1 and males of Fainb3 were placed in 1000-ml beakers filled with medium; every 3 days parthenogenetic offspring were removed to prevent selfing of females with their sons, thereby ensuring that all offspring were outcrossed.
Parasite material
We used spores from P. ramosa clone C1 (Russia, Moscow) for our infection trials, which, as other P. ramosa clones, shows highly specific infectivity (for more details see Luijckx et al., 2011). This parasite clone is both phenotypically and genetically similar to a naturally occurring parasite in Finland (Luijckx et al., 2011; McElroy et al., 2011). Spore suspensions were created by homogenising D. magna with late stage infections in water. Spore concentration in each suspension was determined using a haemocytometer and phase contrast microscopy.
Tests for susceptibility
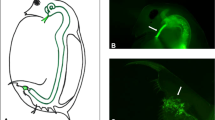
We employed two different techniques to determine host susceptibility: the attachment test and infection trials. The attachment test is a recently developed technique that employs fluorescent microscopy to assess the ability of P. ramosa spores to attach to the oesophagus of Daphnia. Attachment perfectly correlates with susceptibility of the Daphnia host in infection trials (Duneau et al., 2011). Infection trials reflect the outcome of the entire infection process (encounter, attachment, penetration and proliferation within the host), whereas the attachment test only looks at one step in this process, the attachment step. Susceptibility of parental, F1 and F2 Daphnia clones was determined with both methods (numbers of tested clones differed between methods due to loss of some host clones before the end of all tests). Susceptibility in the backcross was determined with the attachment test, with a representative subset of host clones tested with infection trials. Susceptibility of selfed parents was only tested using the attachment test. Both assays agreed very well with each other, although variation between replicates in the infection trails was greater.
Infection trials
To remove maternal effects, mothers of the F1 and F2 were kept singly in 100-ml beakers filled with medium under standard conditions for three asexual generations before the start of the infection trials. For the backcross, this was reduced to one asexual generation. For infection trials, we used female D. magna of 1 to 4 days old at the time of parasite exposure and all were offspring from the third or later clutch. Juveniles were exposed to spores by placing them singly in 100-ml beakers filled with 20 ml of medium and adding spore suspensions containing 200 000 spores of P. ramosa clone C1. This dose is known to cause 100% infections without lethal effect in susceptible hosts (Regoes et al., 2003). Individual D. magna remained in 20 ml of medium for 4 days, at which point the beakers were filled up. For parental clones and the F1, we utilised a split-brood design with eight individuals from different mothers in separate 100-ml beakers with medium under standard conditions for both the treatment and the control. The medium was changed at day 7 of the experiment, and twice weekly for the following 23 days. Daphnia were fed 3 × 105 cells ml−1 algae cells daily throughout the experiment. The F2 clones were tested using the same protocol with the following exceptions. We used four replicates per clone and to accommodate the increasing food demand of the growing animals, feeding was raised from 3 × 105 cells ml−1 to algae 6 × 105 cells ml−1 during the experiment. For the backcross, we tested 40 of the 164 clones using four randomly chosen juveniles (1 to 4 days old) and fed 3 × 105 cells ml−1 algae daily at the start of the experiment and 5 × 105 cells ml−1 towards the end. All infection trials lasted 30 days. Survival was monitored at least once every 48 h. All animals dying after day 12 and those surviving till the end of the experiment were tested for infections by checking for parasite spores using phase-contrast microscopy (magnification × 400). Individuals dying before day 12 of the experiment were not assessed because detection of P. ramosa infection is unreliable during the early stage of infection. Host clones were considered susceptible when one or more replicates were infected and considered resistant when none were infected.
Attachment test
The attachment test is described in full Duneau et al. (2011). In short, for assessment of susceptibility with the attachment test four individuals older than 4 days were taken from stock cultures for each host clone (stocks kept in 100-ml jars and fed 3 × per week with 8 × 107 cells ml−1) and placed singly in 24-well plates in 1 ml of medium. A total of 20 000 fluorescent-labelled spores and contrast dye were added to each well and plates were incubated for 1 h at room temperature. Attachment, indicating susceptibility of the host clone, was determined by examining exposed Daphnia with a fluorescent microscope and checking for the presence of fluorescently labelled spores on the Daphnia oesophagus.
Results
We found a strong binary pattern of resistance; all or all but one replicate of susceptible clones became infected in infection trials, whereas resistant clones never became infected. Three host clones from the backcross were an exception; in these only one of the replicates became infected during the infection trials. The attachment test gave consistent results (with no variance); each host clone displayed either complete resistance or susceptibility.
Parental clones showed contrasting susceptibility, Fainb3 was susceptible, whereas Xinb3 was resistant to P. ramosa C1. Parents were likely homozygous at the relevant loci due to three generations of selfing and indeed offspring of the selfed parents showed identical phenotypes as their parents and no segregation (Figure 1, Table 1). The F1 clone resembled Xinb3 and was resistant. The pattern for resistance in the F2 (56 resistant (79%) and 15 susceptible (21%), combined data from infection trial and attachment test, Table 1), was not significantly different from the 3:1 pattern expected if resistance is determined by a dominant, single-locus trait exhibiting Mendelian segregation (Fisher's exact test P=0.69). The single locus model with dominant inheritance of resistance was confirmed by the backcross in which 84 clones were resistant (51%) and 80 susceptible (49%), which was not significantly different from the expected 50:50 (Fisher's exact test P=0.91, Figure 1, Table 1)
Pedigree showing the Daphnia magna crossing scheme and the resistance/susceptibility profiles to Pasteuria ramosa for parental, F1, backcross and F2 host clones. Black Daphnia indicate susceptibility and unfilled Daphnia indicate resistance. In our proposed model of inheritance, resistance to P. ramosa clone C1 is conferred by a dominant allele, A, and susceptibility by a recessive allele, a.
Discussion
By using two different methods to test for susceptibility, we show that the attachment to the host oesophagus, an important step of the infection process, is controlled by a single host locus. Alleles at this locus segregate in a Mendelian pattern and our crosses revealed the presence of two alleles, a dominant allele ‘A’ preventing attachment of P. ramosa C1 and a recessive allele ‘a’ allowing for attachment (Figure 1). Thus, hosts show binary resistance patterns either being resistant (no attachment) or susceptible (attachment). Attachment to the host oesophagus is responsible for the strong genotype–genotype interactions in the D. magna–P. ramosa system (Duneau et al., 2011; Luijckx et al., 2011). Thus, the finding of a single host locus controlling susceptibility to P. ramosa C1 suggests that these interactions might have a simple genetic basis.
Binary resistance of D. magna clones to P. ramosa clones was found previously (Luijckx et al., 2011) and is related to the way the parasite enters the host (Duneau et al., 2011). During filter feeding spores attach to the oesophagus of susceptible Daphnia and penetrate the oesophagus wall. In resistant Daphnia no attachment is observed. The specificity of attachment is dependent on the genotype of both host and parasite and is not influenced by environmental effects (Duneau et al., 2011). Indeed, we found low variability within Daphnia genotypes in infection trials (all or all but one replicate infected and three cases were only one of four replicates was infected) and no variability in the attachment test, both assays agreed very well with each other. The difference in variance between both tests may be explained by outcome of the attachment test being only determined by the attachment step, whereas the outcome of the infection trials is influenced by the entire infection process, including encounter, attachment and proliferation within the host (Duneau et al., 2011). Contrary to the attachment-step, the entire infection process has been shown to be influenced by environmental (Vale et al., 2008) and maternal effects (Ben-Ami et al., 2010), which may explain the greater variability found in our infection trials. It is likely that other loci other than the one described here are involved in other steps of the infection process. For example, encounter of the parasite spores, which reside in the sediment, may be dependent on diel vertical migration, which has been shown to have a strong genetic component (Decaestecker et al., 2002). It has also been suggested that genes affecting parasite proliferation within the host might be different from those involved in attachment (Decaestecker et al., 2007; Duneau et al., 2011). Even though more loci might be involved in the entire infection process, the locus described here appears to be the major determinant of susceptibility to P. ramosa C1 and is involved in the attachment of the parasite to the host oesophagus, which is a key step in D. magna–P. ramosa co-evolution (Duneau et al., 2011).
Our interpretation that resistance is coded by a single dominant locus is partially consistent with previous studies on the genetic basis of resistance in D. magna. A pervious study speculated that resistance is due to one or a few loci (Little et al., 2006). However, small sample size and use of P. ramosa isolates (not clones) in this earlier study makes comparison difficult. In other invertebrate–parasite systems the majority of resistance genes tend to be dominant and autosomal, examples include: Drosophila-parasitic Wasp (Carton et al., 1992); mosquito-malaria (Thathy et al., 1994); and snail-Schistosoma (Knight et al., 1999; Lewis et al., 2003) (for review of invertebrate resistance see Carton et al., 2005).
P. ramosa clones not tested in this study also require attachment to the host oesophagus for successful infection (Duneau et al., 2011), and all host genotypes tested with P. ramosa clones show binary resistance (Luijckx et al., 2011). Conservation of the infection mechanism among different parasite genotypes and similar phenotypic patterns of susceptibility of the host for other parasite genotypes suggests that our finding of Mendelian inheritance of resistance against P. ramosa may also apply to untested D. magna and P. ramosa genotypes. Resistance to other P. ramosa genotypes may be conferred by additional alleles on the found locus or by additional loci possibly similar to the well-described gene-for-gene resistance in plants (Keen, 1990).
The finding of a simple genetic basis of host resistance has important implications for the outcome of host–parasite co-evolution. Negative frequency-dependent selection may occur in the presence of host–parasite genotype–genotype interactions with simple underlying genetics (Clarke, 1976). Strong genotypic interactions were already described in the Daphnia–Pasteuria system by Luijckx et al. (2011) and their data shows that the locus described here only confers resistance to specific P. ramosa genotypes; the resistant parent (Xinb3) was susceptible to three of the five tested P. ramosa genotypes, whereas other host genotypes were resistant to these P. ramosa. A simple genetic basis for genotype–genotype interactions supports earlier findings of negative frequency-dependent selection for infectivity in the D. magna–P. ramosa system (Decaestecker et al., 2007). Which may be important for the maintenance of genetic variation and the evolution of recombination as suggested by the Red Queen Theory (Jaenike, 1978; Hamilton, 1980).
Many theoretical models used to predict host–parasite co-evolution assume simple genetics with binary resistance patterns (Salathe et al., 2008 and references therein), whereas data from empirical studies suggests that patterns are more quantitative (for example, Schulenburg and Ewbank, 2004). In addition, these models also (often) assume that parasites are highly specific to given host genotypes. We show that in one step of the infection process a single locus is responsible for binary resistance. Furthermore, resistance is highly specific for the here tested P. ramosa genotype. Thus, our results suggests that theoretical models for host–parasite co-evolution may, in some cases, not be over-simplified and hold promise for understanding and interpreting empirical results.
Our results add to the experimental power of the D. magna–P. ramosa model system as a tool for understanding the evolution of host–parasite interactions. Furthermore, the isolation of more P. ramosa clones and additional crosses between host clones will allow for the creation of a D. magna–P. ramosa interaction matrix in which infectivity profiles can be determined by pair-wise matching of D. magna and P. ramosa genotypes. Through competition experiments, use of natural populations and the development of a Quantitative Trait Locus panel (Routtu et al., 2010), this matrix has the possibility to serve as a powerful tool for testing evolutionary models.
Data archiving
Data have been deposited at Dryad: doi:10.5061/dryad.6j1qv1m3
References
Ben-Ami F, Ebert D, Regoes RR (2010). Pathogen dose infectivity curves as a method to analyze the distribution of host susceptibility: a quantitative assessment of maternal effects after food stress and pathogen exposure. Am Nat 175: 106–115.
Ben-Ami F, Regoes RR, Ebert D (2008). A quantitative test of the relationship between parasite dose and infection probability across different host-parasite combinations. Proc R Soc B Biol Sci 275: 853–859.
Carius HJ, Little TJ, Ebert D (2001). Genetic variation in a host-parasite association: potential for coevolution and frequency-dependent selection. Evolution 55: 1136–1145.
Carton Y, Frey F, Nappi A (1992). Genetic determinism of the cellular immune-reaction in Drosophila melanogaster. Heredity 69: 393–399.
Carton Y, Nappi AJ, Poirie M (2005). Genetics of anti-parasite resistance in invertebrates. Dev Comp Immunol 29: 9–32.
Clarke B (1976). The ecological genetics of host-parasite relationships. In: Taylor AER, Muller R (eds). Genetic Aspects of Host-Parasite Relationships. Blackwell: Oxford, pp 87–103.
Colson I, Du Pasquier L, Ebert D (2009). Intragenic tandem repeats in Daphnia magna: structure, function and distribution. BMC Res Notes 2: 206.
Decaestecker E, De Meester L, Ebert D (2002). In deep trouble: Habitat selection constrained by multiple enemies in zooplankton. Proc Natl Acad Sci USA 99: 5481–5485.
Decaestecker E, Gaba S, Raeymaekers JAM, Stoks R, Van Kerckhoven L, Ebert D et al. (2007). Host-parasite ‘Red Queen’ dynamics archived in pond sediment. Nature 450: 870–873.
Duneau D, Luijckx P, Ben-Ami F, Laforsch C, Ebert D (2011). Resolving the infection process reveals striking differences in the contribution of phylogeny, genetics and environment to host-parasite interactions. BMC Biol 9: 11.
Ebert D, Carius HJ, Little T, Decaestecker E (2004). The evolution of virulence when parasites cause host castration and gigantism. Am Nat 164: S19–S32.
Ebert D, Rainey P, Embley TM, Scholz D (1996). Development, life cycle, ultrastructure and phylogenetic position of Pasteuria ramosa Metchnikoff 1888: rediscovery of an obligate endoparasite of Daphnia magna Straus. Philos Trans R Soc Lond Ser B Biol Sci 351: 1689–1701.
Ebert D, Zschokke-Rohringer CD, Carius HJ (1998). Within- and between-population variation for resistance of Daphnia magna to the bacterial endoparasite Pasteuria ramosa. Proc R Soc Lond Ser B Biol Sci 265: 2127–2134.
Grigg ME, Suzuki Y (2003). Sexual recombination and clonal evolution of virulence in Toxoplasma. Microbes Infect 5: 685–690.
Hamilton WD (1980). Sex versus non-sex versus parasite. Oikos 35: 282–290.
Henter HJ, Via S (1995). The potential for coevolution in a host-parasitoid system.1. Genetic-variation within an aphid population in susceptibility to a parasitic wasp. Evolution 49: 427–438.
Jaenike J (1978). An hypothesis to account for the maintenance of sex within populations. Evolutionary Theory 3: 191–194.
Jensen KH, Little T, Skorping A, Ebert D (2006). Empirical support for optimal virulence in a castrating parasite. Plos Biol 4: 1265–1269.
Keen NT (1990). Gene-for-gene complementarity in plant-pathogen interactions. Annu Rev Genet 24: 447–463.
Knight M, Miller AN, Patterson Cn, Rowe CG, Michaels G, Carr D et al. (1999). The identification of markers segregating with resistance to Schistosoma mansoni infection in the snail Biomphalaria glabrata. Proc Natl Acad Sci USA 96: 1510–1515.
Lambrechts L, Halbert J, Durand P, Gouagna LC, Koella JC (2005). Host genotype by parasite genotype interactions underlying the resistance of anopheline mosquitoes to Plasmodium falciparum. Malaria J 4: 3.
Lewis FA, Patterson CN, Grzywacz C (2003). Parasite-susceptibility phenotypes of F-1 Biomphalaria glabrata progeny derived from interbreeding Schistosoma mansoni-resistant and -susceptible snails. Parasitol Res 89: 98–101.
Little TJ, Ebert D (2000). The cause of parasitic infection in natural populations of Daphnia (Crustacea : Cladocera): the role of host genetics. Proc R Soc Lond Ser B Biol Sci 267: 2037–2042.
Little TJ, Watt K, Ebert D (2006). Parasite-host specificity: experimental studies on the basis of parasite adaptation. Evolution 60: 31–38.
Lively CM, Dybdahl MF (2000). Parasite adaptation to locally common host genotypes. Nature 405: 679–681.
Luijckx P, Ben-Ami F, Mouton L, Du Pasquier L, Ebert D (2011). Cloning of the unculturable parasite Pasteuria ramosa and its Daphnia host reveals extreme genotype-genotype interactions. Ecol Letters 14: 125–131.
McElroy K, Mouton L, Du Pasquier L, Qi W, Ebert D (2011). Characterisation of a large family of polymorphic collagen-like proteins in the endospore-forming bacterium Pasteuria ramosa. Res Microbiol 162: 701–714.
Morand S, Manning SD, Woolhouse MEJ (1996). Parasite-host coevolution and geographic patterns of parasite infectivity and host susceptibility. Proc R Soc Lond Ser B Biol Sci 263: 119–128.
Mouton L, Nong G, Preston JF, Ebert D (2007). Variable-number tandem repeats as molecular markers for biotypes of Pasteuria ramosa in Daphnia spp. Appl Environ Microbiol 73: 3715–3718.
Nowak MA, May RM (1994). Superinfection and the evolution of parasite virulence. Proc R Soc Lond Ser B Biol Sci 255: 81–89.
Read AF, Taylor LH (2001). The ecology of genetically diverse infections. Science 292: 1099–1102.
Regoes RR, Hottinger JW, Sygnarski L, Ebert D (2003). The infection rate of Daphnia magna by Pasteuria ramosa conforms with the mass-action principle. Epidemiol Infect 131: 957–966.
Routtu J, Jansen B, Colson I, De Meester L, Ebert D (2010). The first-generation Daphnia magna linkage map. BMC Genomics 11: 508.
Salathe M, Kouyos RD, Bonhoeffer S (2008). The state of affairs in the kingdom of the Red Queen. Trends Ecol Evol 23: 439–445.
Schmid-Hempel P, Funk CR (2004). The distribution of genotypes of the trypanosome parasite, Crithidia bombi, in populations of its host, Bombus terrestris. Parasitology 129: 147–158.
Schmid-Hempel P, Puhr K, Kruger N, Reber C, Schmid-Hempel R (1999). Dynamic and genetic consequences of variation in horizontal transmission for a microparasitic infection. Evolution 53: 426–434.
Schulenburg H, Ewbank JJ (2004). Diversity and specificity in the interaction between Caenorhabditis elegans and the pathogen Serratia marcescens. BMC Evol Biol 4: 49.
Thathy V, Severson DW, Christensen BM (1994). Reinterpretation of the genetics of susceptibility of Aedes aegypti to Plasmodium gallinaceum. J Parasitol 80: 705–712.
Thompson JN, Burdon JJ (1992). Gene-for-gene coevolution between plants and parasites. Nature 360: 121–125.
Vale PF, Stjernman M, Little TJ (2008). Temperature-dependent costs of parasitism and maintenance of polymorphism under genotype-by-environment interactions. J Evol Biol 21: 1418–1427.
Webster JP, Woolhouse MEJ (1998). Selection and strain specificity of compatibility between snail intermediate hosts and their parasitic schistosomes. Evolution 52: 1627–1634.
Wilfert L, Gadau J, Baer B, Schmid-Hempel P (2007). Natural variation in the genetic architecture of a host-parasite interaction in the bumblebee Bombus terrestris. Mol Ecol 16: 1327–1339.
Acknowledgements
We thank J Hottinger, N Boileau and U Stiefel for assistance in the laboratory. This study was supported by the Swiss National Science Foundation. Harris Fienberg was supported by a Fulbright grant.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Rights and permissions
About this article
Cite this article
Luijckx, P., Fienberg, H., Duneau, D. et al. Resistance to a bacterial parasite in the crustacean Daphnia magna shows Mendelian segregation with dominance. Heredity 108, 547–551 (2012). https://doi.org/10.1038/hdy.2011.122
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/hdy.2011.122
Keywords
This article is cited by
-
A SNP assay for assessing diversity in immune genes in the honey bee (Apis mellifera L.)
Scientific Reports (2021)
-
An alternative route of bacterial infection associated with a novel resistance locus in the Daphnia–Pasteuria host–parasite system
Heredity (2020)
-
Host-parasite coevolution in populations of constant and variable size
BMC Evolutionary Biology (2015)
-
Genetic architecture of resistance in Daphnia hosts against two species of host-specific parasites
Heredity (2015)
-
Water fleas require microbiota for survival, growth and reproduction
The ISME Journal (2015)