Abstract
The estimation of recent gene immigration is fundamental to a wide range of evolutionary and conservation studies. In a risk assessment context, gene flow estimation procedures are needed that are both accurate and readily amenable to formal evaluation of statistical uncertainty. However, genetic methods for estimating recent migration rates that are specific and have been thoroughly evaluated are scarce. Here we use an original and straightforward maximum-likelihood method to estimate recent uniparental gametic immigration from non-local plantations into an endangered population of the Iberian relict pine variety Pinus sylvestris var. nevadensis D. H. Christ. Our approach is not intended to ascertain population membership of individuals, but rather to obtain accurate immigration rate estimates with reliable confidence limits. We found very high (∼40%) pollen introgression at the seed-crop level into the Scots pine relict, and substantial (10–15%) male gametic introgression among naturally regenerated recruits. Using numerical simulation, we show that our method yields uniparental gametic immigration estimates that are expected to be virtually unbiased and usually accurate under our sampling conditions. Among four tested methods to estimate the confidence intervals for immigration estimates, the profile-likelihood method was the best, as it outperformed bootstrapping procedures and yielded coverage close to nominal limits under different sample sizes and migration rates. This study presents a method by which researchers can facilitate decision making within a gene flow risk assessment context.
Similar content being viewed by others
Introduction
Gene flow has contrasting implications for conservation biology. On the one hand, it is thought to hamper local adaptation in heterogeneous environments by disrupting co-adapted gene complexes and changing allele frequencies in a direction opposite to divergent natural selection (‘migration load’ effect). On the other hand, gene flow can reduce the deleterious effects of inbreeding and increase the genotypic variance available for selection, facilitating local adaptation (‘genetic rescue’ effect). Although the relative importance of these effects remains unclear, the precise level of gene flow seems critical in determining the final outcome on local adaptation, for a given demographic and selective scenario (Lenormand, 2002). In practice, any objective decision regarding management of ongoing gene flow into conservation populations should be based on accurate estimates of the realized gene immigration rates, which must be appraised along with the specific demographic and selective processes affecting the population.
Estimating gene immigration rates is especially relevant when dealing with protected populations that are potentially exposed to gene flow from artificially introduced interfertile relatives. Any scientific risk assessment protocol of non-local germplasm will require reliable contemporary gene flow estimates, sometimes over broad spatial scales, which may not be easily available. Difficulties in obtaining precise estimates of ongoing gene flow at a landscape scale are illustrated by the intense scientific debate about the presence or absence of gene flow from transgenic plantations into maize landraces in Mexico: more than 10 studies conducted in recent years have disputed various statistical and sampling issues (Mercer and Wainwright, 2008). Many of the apparent inconsistencies among these studies are most likely due to insufficient evaluation of estimation errors. Establishing the statistical uncertainty of gene flow estimates is as important as minimizing their bias for incorporating scientific advice into decision making. However, this aspect of estimation is frequently neglected.
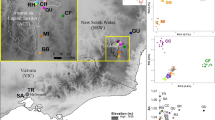
We consider here the illustrative example of Scots pine (Pinus sylvestris L.) populations in Mediterranean Spain. Pinus sylvestris var. nevadensis D. H. Christ is one of the most widely recognized varieties (Farjon, 1998) of this monoecious, wind-pollinated, predominantly outcrossing conifer. It is restricted to a few relict populations in southern Spain, comprising the Trevenque population, which is scattered over 120 hectares at the southernmost extremity of the pine's range, within the Sierra Nevada National Park. This area is closely surrounded by several thousand hectares of conspecific tree plantations not belonging to the nevadensis variety (Figure 1). Introgression from the plantations is a current matter of concern for the National Park managers and local foresters, because mtDNA (Sinclair et al., 1999), cpDNA (Provan et al., 1998) and quantitative growth and survival traits (Alía et al., 2001) have shown a substantial genetic divergence of P. sylvestris var. nevadensis relative to other Iberian and European populations of the species.
Male gametic introgression rates (in black) from non-local plantations into the natural regeneration plots of Pinus sylvestris L. var. nevadensis relict population at Trevenque (Sierra Nevada, Spain). Lines delimit the natural stands, with all the observable surrounding forests being non-local conspecific plantations. The seven southernmost plots are in a dense area (REG1, named ‘La Cortijuela’), whereas the remaining nine are within sparser stands growing on a different slope (REG2, named ‘La Dehesilla’). The estimates significantly different from 0 (95% confidence interval not containing 0) are indicated by * (if only direct estimation of haplotype frequencies yielded a positive test) or by ** (if both direct and Bayesian estimation of haplotype frequencies yielded positive tests).
Few formal procedures are available for estimating contemporary seed and pollen migration over broad spatial scales, as required to assess genetic introgression into the Scots pine relict. Genetic parentage exclusion methods have been developed for assessing immigration into small populations, such as seed orchards (Adams et al., 1997; Plomion et al., 2001; Stoehr and Newton, 2002; Slavov et al., 2005), but these methods are not feasible for large-scale experiments, because they require exhaustive genotyping of all potential parents within the recipient population. Genetic assignment methods, on the other hand, have been designed primarily to ascertain population membership of individuals and admixture proportions, and are not suitable for addressing the statistical question of obtaining accurate migration rates exclusively (Manel et al., 2005). Although they can be used to obtain rough migration rate estimates (Manel et al., 2005), assignment methods typically offer low power when candidate populations are genetically close and do not account for cryptic immigration (immigrants wrongly identified as non-immigrants) when deriving migration rate estimates (Cornuet et al., 1999; Paetkau et al., 2004).
The Bayesian approaches of Wilson and Rannala (2003) and Faubet and Gaggiotti (2008) estimate posterior probability distributions of several population parameters, including recent migration rates among populations. Although these methods are potentially useful for conservation plans requiring recent genetic immigration measures, their accuracy and the reliability of the associated credibility intervals have not been tested under varied realistic demographic conditions. This is not an easy task to perform numerically, given the slowness and frequent convergence problems of the Bayesian estimation algorithms (Faubet et al., 2007; Faubet and Gaggiotti, 2008), which limit their utility within a risk assessment context. In addition, these advanced procedures use diploid biparentally inherited markers and are designed to address the general problem of jointly estimating gene migration rates, population frequencies, inbreeding coefficients and the origin of every individual in the sample. We lack more specific and efficient approaches to estimate migration rates exclusively, for which we could exploit the methodological and inferential advantages of haploid uniparentally inherited DNA markers.
Here we use a straightforward maximum-likelihood (ML) procedure to estimate recent male gametic immigration rates based on paternally inherited chloroplast DNA markers. We assume a single external source of potential immigrants, albeit extension to more than one source population is straightforward. Unlike parentage exclusion methods, our approach does not require exhaustive genotyping of the recipient population. As opposed to assignment methods (Paetkau et al., 1995, 2004; Rannala and Mountain, 1997; Cornuet et al., 1999; Pella and Masuda, 2001; Wilson and Rannala, 2003; Faubet and Gaggiotti, 2008), here we do not aim at establishing population membership of every individual in the sample, but only to obtain an accurate immigration rate estimate and place reliable confidence limits around it. We apply this method to estimate allochthonous male gametic immigration into the Scots pine relict population in the Sierra Nevada National Park, showing how researchers can improve decision making within a gene flow risk assessment context.
Materials and methods
Field sampling
Sampling was conducted in three stages. We first collected needle tissue from nN=112 and nP=108 adult individuals from the native Pinus sylvestris var. nevadensis population (N) and the allochthonous plantations (P), respectively, which we used to select discriminating markers and estimate adult haplotypic frequencies. Adult trees from N were more than a century old and were naturally established well before P was introduced about 50 years ago. We distinguished two sampling areas within the natural population: a dense area (N1, locally known as ‘La Cortijuela’; nN1=63) and a sparser one on a different slope (N2, locally known as ‘La Dehesilla’; nN2=49) (Figure 1). We extracted total DNA and screened all 220 adult trees at six chloroplast microsatellite markers (cpSSR)—Pt1254, Pt15169, Pt26081, Pt30204, Pt36480 and Pt87268 (Vendramin et al., 1996)—according to the procedures detailed by Robledo-Arnuncio et al. (2005). After the initial screening, we identified four markers (Pt15169, Pt26081, Pt36480 and Pt87268) that provided most of the discriminating power between adult individuals of N and P, including some discriminant size variants that were present only in the plantations.
Second, in spring 2004, we sampled a total of 325 recruits (0–30 years old) from 16 natural regeneration plots distributed under the canopy of N (Figure 1). We used this sample to estimate the male gametic introgression rate (m) at the level of regeneration, which may occur either through seed dispersal from N mothers that have been pollinated by P fathers or by seed dispersal from P mothers pollinated by P fathers. Reproductive recruits were very rare and we considered that no backcrosses had taken place in the population. Our sampling estimated the accumulated proportion of immigrant male gametes arriving from the plantations from the time planted trees reached maturity (about 30 years ago) to the present. Seven of the plots (nS=138 individuals) were in the dense N1 area (REG-1), and the other nine plots (nS=187 individuals) were within the sparser N2 area (REG-2). The mean number of sampled recruits per plot was 20.3 (s.d.=6.0). Each of the 325 samples from the natural regeneration was scored at the same four cpSSR that adult individuals were scored at.
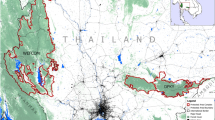
Finally, in winter 2004, we collected a total of nS=440 seeds from 22 seed trees at 11 sampling locations within the REG-1 area (one pair at each location), with a fixed number of 40 seeds per tree pair (Figure 2). We used this sample to estimate male gametic introgression (possible only through pollen in this case) from P into the standing seed crop of N over the pollen dispersal episode of the year 2003. Seeds were germinated and the embryo of each seed was characterized with the selected cpSSR.
Pollen immigration rates (in black) from non-local plantations into the standing seed crop of Scots pine relict population at ‘La Cortijuela’, Trevenque. The estimates significantly different from 0 (95% confidence interval not containing 0) are indicated by * (if only direct estimation of haplotype frequencies yielded a positive test) or by ** (if both direct and Bayesian estimation of haplotype frequencies yielded positive tests).
Model for estimating immigration rates
We defined recent male gametic immigration rate (m) as the proportion of male gametes arriving at N from P during the reference migration period. In our study at Trevenque, the reference period was the previous 30 years (since the plantations reached maturity) for naturally regenerating seedlings and the 2003 pollination episode for the seed crop collected in 2004. Consider a sample S of nS individuals from population N, collected after the reference migration period, for which we want to know m. This would be either the sample of seeds or naturally regenerating seedlings. Let k be the number of chloroplast haplotypes (cp-haplotypes), defined as unique combinations of variants at the set of chloroplast DNA marker regions, and let qh,N and qh,P be the frequencies before the reference migration episode of the h-th haplotype in populations N and P, respectively. Under random mating, the probability that cp-haplotype h is observed in the sample S (after migration) is

In the favourable case that there was a set of l plantation-specific cp-haplotypes (h=1, 2,…, l with qh,N=0), a direct estimator of the male gametic immigration rate would then be

where qh,S is the frequency of the h-th cp-haplotype in the sample S. More generally, we can use equation (1) to compute the joint-likelihood function for the whole set of cp-haplotypes (that is, including both plantation-specific and shared ones) carried by the nS individuals in the sample S as a function of m:

and estimate m by maximizing the log-likelihood function

Note that the estimator of m obtained from equation (4) accounts for cryptic immigration, as it defines the probability of observing a cp-haplotype in the recipient population as a function of m and its frequency in both the candidate source (qh,P) and recipient (qh,N) populations. On the basis of the estimated adult population cp-haplotype frequencies (see section ‘Estimating population haplotype frequencies’), we used either equation (2) or (4) to obtain two estimates of m at each of the two natural regeneration areas REG-1 and REG-2 (nS=138 and 187 seedling cp-haplotypes, respectively) and at the seed-crop level (nS=440 seed embryo cp-haplotypes). In order to show potential spatial heterogeneities in introgression levels, we also estimated m separately at each of the 16 natural regeneration plots and for each of the 40 embryo samples collected from each seed-tree pair.
Estimating population haplotype frequencies
The population cp-haplotype frequencies before migration (the qh,N−s and the qh,P−s) must be estimated from the field samples. Let xh,N and xh,P be the observed counts of the h-th cp-haplotype in the adult samples from populations N and P, respectively (with h=1, 2,…, k). Maximum-likelihood cp-haplotype frequency estimates are given by

If the sample sizes nN and nP were very large, we might safely assume that the cp-haplotype frequencies estimated using equation (5) are close enough to the exact population values, and use them in equation (4) to obtain an immigration estimate m̂. In a decision-making context, however, it may be appropriate to minimize the type I error (non-immigrants wrongly identified as immigrants) in order to avoid potential overestimates of m, which could motivate unnecessary and costly actions to reduce introgression. A source of type I error could be the presence of low-frequency plantation-specific cp-haplotypes in the observed sample (q̂h,N=0 and a small q̂h,P>0), if in fact they are present (but not detected) in the natural population (small qh,N>0). We could then assume a priori that all cp-haplotypes are shared between P and N at equal frequency, and estimate haplotype population frequencies on the basis of this prior assumption and the observed counts. For this purpose, we use the Bayesian approach described by Rannala and Mountain (1997), which gives posterior cp-haplotype frequencies as

where K is the total number of cp-haplotypes in the two populations, which we estimate with the number (k) of observed cp-haplotypes in the sample. By using equation (4) and the cp-haplotype frequencies estimated with equation (6), we obtain a conservative immigration rate estimate (m̂′) accounting for potential overestimates derived from undetected low-frequency cp-haplotypes in the recipient population.
Assessing statistical uncertainty
Given the two vectors of estimated cp-haplotype frequencies for populations N {q̂1,N, q̂2,N, …, q̂k,N} and P {q̂1,P, q̂2,P, …, q̂k,P}, we computed the expected bias and accuracy (root mean square error, RMSE) of m̂ for a sample size nS and an assumed introgression rate m by simulating stochastic samples of size nS as follows: (i) draw a random number r in the half-closed interval [0, 1); (ii) generate an individual i with assigned source population P if r<m, or with assigned source population N otherwise; (iii) randomly draw a cp-haplotype for individual i from a multinomial distribution with k possible outcomes, with respective probabilities {q̂1,N, q̂2,N, …, q̂k,N} if i comes from N, or with respective probabilities {q̂1,P, q̂2,P, …, q̂k,P} if i comes from P; (iv) start again from the first step until obtaining nS individuals. Next, we compute m̂ by applying equation (2) (or equation (4)) to the simulated sample of size nS and repeat the whole procedure to generate 10 000 independent random samples with their corresponding m̂ values. On the basis of the 10 000 m̂ values, we compute the expected relative bias and expected relative RMSE of the m estimator for the assumed sample size, migration rate and cp-haplotype frequencies.
Next, we assessed the performance of four common methods to estimate confidence intervals (CIs): the standard bootstrap (S-Boot), the simple percentile bootstrap (SP-Boot), the accelerated bias-corrected percentile bootstrap (ABC-Boot) and the profile-likelihood method (P-Likelihood). S-Boot is based on variance estimates and the assumption of normality for the distribution of the estimator, whereas SP-Boot is free of this assumption. ABC-Boot accounts for potential biases arising from skewed bootstrap distributions. Finally, P-Likelihood is based on the inspection of the profile-likelihood function. More detailed descriptions of these statistical procedures can be found elsewhere (for example, Manly, 1997, pp 34–55; Coles, 2001, pp 34–35). On the basis of the simulated data obtained with the Monte Carlo approach described above, we estimated 95% CIs for m̂, one for each of the 10 000 independent samples, using these four different methods. Next, we computed the coverage of each method as the proportion of the 10 000 estimated CIs that contained the assumed immigration estimate (m) in the simulations (Manly, 1997). Specifically, we assessed the proportion of times that the upper confidence limit was too low (smaller than m) and the proportion of times that the lower confidence limit was too high (larger than m), comparing these proportions to their nominal 2.5% value.
In the simulations, we assumed that haplotype population frequencies were equal to the ML frequencies (equation (5)), and we considered four sample sizes: nS=20, 40, 138 and 440, corresponding to the real sample sizes used in our empirical study. For each sample size, we assumed migration rates (m) ranging from 0 to 100%. We did not intend to evaluate the statistical behaviour of the methods under a broad range of sampling, genetic differentiation and immigration conditions, but rather to exemplify the assessment of the statistical uncertainty of introgression estimates for a particular data set in a real decision-making scenario.
Results
Genetic diversity and haplotype frequencies
Haplotype richness among adult trees was substantially lower in the Scots pine relict population (N) than in the surrounding plantations (P), with 7 and 24 observed cp-haplotypes, respectively (Table 1). The effective number of cp-haplotypes, calculated as the inverse of the unbiased haplotypic diversity, was 2.8 in N and 7.4 in P. Haplotypic differentiation between N (N1 and N2 pooled together) and P was FST=0.048 (P<0.01), whereas that between N1 and N2 was FST=0.034 (P=0.041). Although all cp-haplotypes found in N were present in P, 17 putative plantation-specific cp-haplotypes were detected at low individual (q̂h,P=0.009–0.0463) but substantial cumulative (∑q̂h,P=0.278) frequencies (Table 1). Under the prior assumption that all haplotypes are shared between N and P, individual posterior frequencies in the natural stands for putatively plantation-specific cp-haplotypes were low: q̂′h,N1=0.0007 for N1 and q̂′h,N2=0.0008 for N2 (Table 1).
Most of the 325 naturally regenerated recruits collected below the canopy of N showed the same set of seven cp-haplotypes shared among the adults of N and P, but a few of the putatively plantation-specific cp-haplotypes were also detected at low frequency among the recruits (Table 1). The observed cumulative frequencies of putatively plantation-specific cp-haplotypes were 0.029 at REG-1 and 0.0374 at REG-2. Among the 440 seeds collected from the standing crop of adult trees of N, a total of 387 carried cp-haplotypes shared between adult trees from N and P (Table 1), whereas 30 seeds carried putative plantation-specific cp-haplotypes (with a cumulative frequency of 0.068; Table 1), and the remaining 23 seeds showed 23 different cp-haplotypes that were absent among sampled adult trees of both N and P (data not shown).
The 23 rare haplotypes (frequency=0.0023) found in the seed crop were probably present in the adult populations N and P but remained undetected because of the smaller adult sample sizes. An alternative, yet unlikely, explanation is that some haplotypes might have dispersed from distant populations (the closest conspecific population is more than 100 km away) or may have arisen by mutation or genotyping error. In either case, as the estimated frequencies for these 23 haplotypes are identical among adult trees of N and P (0 if estimated directly or a small value close to 0 if estimated using the Bayesian approach), they lack inferential value for the estimation of male gametic immigration from P into N, so we discarded them from subsequent analysis.
Immigration estimates
The estimates of male gametic immigration (m) from P into N did not differ significantly when considering adult tree frequencies of N1 and N2 pooled together or separately (results not shown), so we only report here estimates based on the latter. When only putatively plantation-specific cp-haplotypes were used (equation (2)), estimated immigration rates of male gametes from the plantation into the natural population were m̂=0.029/0.2778=0.104 for REG-1, m̂=0.0374/0.2778=0.135 for REG-2 and m̂=0.068/0.2778=0.245 for the seed crop. On the basis of both plantation-specific and shared cp-haplotypes (equation (4)), estimates of male gamete immigration when directly estimating adult haplotype frequencies with equation (5) were m̂=0.143 (REG-1), 0.121 (REG-2) and 0.387 (seed crop), whereas the corresponding values under the prior assumption that all cp-haplotypes are shared between the natural population and the plantation (equation (6)) were m̂=0.116 (REG-1), 0.044 (REG-2) and 0.369 (seed crop) (Table 2).
Immigration estimates at the regeneration-plot level ranged from 0 to 45%, and did not reveal a clear spatial pattern; plots showing larger values appear scattered over the whole area (Figure 1). Pollen immigration at the seed-crop level ranged from 7 to 70% among seed trees, and in this case, progenies showing the highest immigration rates were generally those from seed trees located in narrow elongated areas or small fragments embedded within the plantations (Figure 2).
Expected bias and accuracy
Simulation results indicated that m-estimates exclusively based on plantation-specific cp-haplotypes (equation (2)) had as low a bias as ML estimates based on the full set of observed haplotypes (equation (4)), but were consistently less accurate (relative RMSE 5–80% larger) for all sample sizes and assumed immigration rates (results not shown). The sensitivities of the two estimators to both sample size and assumed immigration rate were qualitatively and quantitatively very similar. Therefore, here we comment only on the results corresponding to the more accurate ML approach.
The ML estimation method (equation (4)) is expected to yield male gametic immigration estimates with very low bias for all nS and m values considered (Table 3). The expected relative bias was smaller than 6% in all cases, and smaller than 3% in most cases and did not show a constant sign across simulation conditions. The expected relative RMSE was sensitive to both nS and m, with a marked decreasing trend with increasing nS and m. This trend was determined by the higher variance of the estimator for small sample sizes and small parametric m values (MSE=Bias2+Variance). The relative RMSE reached a high of 1.8 for nS=20 and m=0.05, whereas it remained below 0.4 for any value of m when nS=440 (Table 3). Considering the real sample sizes and estimated immigration rates for Scots pine, the ML estimator is expected to be virtually unbiased in all cases, while its expected relative RMSE would be about 0.4 for REG-1 (nS=138, m̂=0.143), 0.35 for REG-2 (nS=187, m̂=0.121) and 0.1 for the seed crop (nS=440, m̂=0.387). The expected relative RMSE for immigration estimates at the regeneration plot (nS=20) and seed-tree pair (nS=40) levels are more variable and generally higher, ranging from 0.2 to 1.8 for different values of m (Table 3).
Confidence interval estimation
Simulations indicate that the four tested methods for finding CIs for m-estimates yield considerably different coverage for constant 95% nominal limits, and that all methods generally perform better for larger nS and m (Table 4). Overall, P-Likelihood clearly gave the best and most robust performance, with its limits including the parametric m between 91 and 100% of the time across the range of assumed nS and m values. Both the upper and lower limits calculated by P-Likelihood were quite satisfactory, ranging from 0 to 5%, close to the nominal 2.5%. By contrast, the three bootstrapping approaches gave generally bad performances and were far more sensitive to nS and m. For nS<138 and m<0.1, all S-Boot, SP-Boot and ABC-Boot methods yielded CIs including the parametric m as infrequently as 65% of the time, with their upper limit being too low 20–30% of the time. For larger sample size (nS=440), all four methods tended to give similar performances, especially when m>0.2, with SP-Boot very slightly outperforming P-Likelihood for m=0.4 (Table 4).
On the basis of the simulation results, we chose P-Likelihood for estimating CIs for field estimates of m at the regeneration-plot and seed-tree pair levels, as well as for the global natural regeneration estimates. We restricted the use of SP-Boot for estimating the CI of m̂ for the global seed-crop sample. The results indicate that the global male gametic immigration estimates are significantly different from 0 both at the regeneration (REG-1 and REG-2) and seed-crop level when using ML estimates of cp-haplotype frequencies (Table 2). However, considering the conservative prior assumption that the natural population and the plantation share all cp-haplotypes, we cannot reject the hypothesis that the global immigration rate is 0 at REG-2 (Table 2). On a finer scale, only the largest estimates were significantly different from 0 at the regeneration-plot level, but virtually all estimates were significantly different from 0 at the seed-tree pair level (Figures 1 and 2).
Discussion
We used a novel and straightforward ML method, on the basis of haploid uniparentally inherited genetic markers, to estimate recent uniparental gametic immigration from non-local plantations into a relict Scots pine population. By exclusively estimating immigration rates, we circumvented convergence issues, large computation time and potential biases associated with more general methods involving the joint estimation of migration rates, population membership of individuals, allelic frequencies and other population coefficients (Pella and Masuda, 2001; Wilson and Rannala, 2003; Faubet et al., 2007; Faubet and Gaggiotti, 2008). The Monte Carlo simulation results show that the proposed method can be expected to yield virtually unbiased and fairly accurate uniparental gametic immigration estimates under our sampling conditions. In addition, we have illustrated how our approach fits within a gene flow risk assessment framework, allowing a reliable evaluation of statistical uncertainty of immigration estimates.
Our definition of recent migration refers to the exchange of migrants occurred during a reference migration period, such as episodes of pollen and seed dispersal, before and after which we can unambiguously estimate haplotype population frequencies. This sequential sampling approach, feasible for organisms with discrete synchronized migration periods, avoids the necessary joint estimation of allele frequencies (before migration) of methods that compute migration rates using the target sample of potential immigrants also as reference set (Wilson and Rannala, 2003; Faubet and Gaggiotti, 2008).
The estimation of uniparental gametic immigration using haploid uniparentally inherited markers represents a more tractable problem than obtaining total immigration rates from diploid biparentally inherited genetic data. We neither required assuming Hardy–Weinberg proportions or linkage equilibrium among loci nor required jointly estimating inbreeding coefficients. Consequently, the simplicity of the log-likelihood function (equation (4)) allows an extremely fast optimization that facilitates the evaluation of the robustness and accuracy of the estimator. As pointed out by Wilson and Rannala (2003), it is advisable to evaluate numerically the reliability of migration rate estimates for particular data sets, given the observed levels of genetic differentiation. However, assessing the statistical properties of an estimator on the basis of as few as 10 replicates, a frequently adopted limit imposed by the time-consuming available Bayesian methods to estimate recent migration (Wilson and Rannala, 2003; Faubet et al., 2007; Faubet and Gaggiotti, 2008) may be insufficient (Manly, 1997). This limitation can be a serious drawback in real risk-assessment scenarios. By contrast, we were able to conduct heavily (10 000) replicated simulations in our case study, which provides a sounder basis for calculating the expected accuracy of migration estimates and choosing a reliable method for deriving CIs. Indeed, as suggested by our simulation results, uninformed method selection for assessing statistical uncertainty could yield flawed and misleading CIs for migration rates.
Our numerical analyses exemplify the assessment of the statistical uncertainty of introgression estimates for a particular data set in a real decision-making scenario. Future theoretical studies should evaluate the statistical behaviour and potential limitations of the proposed estimation method under a broad range of sampling, demographic and genetic differentiation conditions. Of particular interest will be how the accuracy of immigration estimates and uncertainty measures are affected by very low levels of genetic differentiation (FST≪0.05) among immigrant sources and/or by the presence of multiple (sampled or unsampled) potential source populations.
The possibility of obtaining uniparental gametic immigration measures on a landscape scale is relevant to many scientific and practical problems, such as evolutionary investigations about the consequences of sex-biased dispersal among populations (Hu and Ennos, 1999; Lopez et al., 2008) or conservation studies evaluating the relative exposure of natural plant populations to seed and pollen from exotic or transgenic plantations. When using paternally inherited markers, as in the Scots pine case study, migration rate estimates will measure pollen immigration only if the target sample is a set of seeds collected before dispersal, whereas estimates will reflect immigration of male gametes by both pollen and seed dispersal if the sample is a set of individuals collected after seed dispersal. Alternatively, given that the only possible vector for female gametic immigration is seed dispersal, estimates obtained using haploid maternally inherited markers would necessarily measure seed immigration rates (note that our approach could also be used for this purpose). By combining genetic markers with different modes of uniparental inheritance, it would be possible to estimate ratios of male to female gamete immigration rates.
The empirical results from this study reveal some considerations for conservation management of the Scots pine relict population at Trevenque. Our estimates indicate very high (about 40%) pollen introgression from non-local plantations at the seed-crop level before seed dispersal and substantial (10–15%) male gametic introgression among naturally regenerated recruits, both being significantly different from 0. If managers chose to work under the conservative prior assumption that all haplotypes are shared between the populations, however, we could not conclude that the introgression levels are significantly different from 0 among recruits sampled in the ‘La Dehesilla’ area (REG-2), which, based on this evidence, could be considered a candidate for a gene reserve.
Interestingly, male gametic immigration was about threefold higher at the seed-crop level than at the regeneration level (P=0.001; from 105 random permutations of individuals between samples). Taking a closer look at the regeneration sample, we also observed a decreasing trend of immigration rates with recruit age: 0.300 (95% CI: 0.117–0.585) for <10 years (nS=46), 0.189 (95% CI: 0.112–0.290) for 10–20 years (nS=214) and 0.113 (95% CI: 0.008–0.416) for 20–30 years (nS=30), although these estimates were not significantly different from each other (P>0.1). Two main hypotheses with contrasting management implications can be proposed to explain these trends: (i) immigrants sired by pollen donors from the plantations have a post-dispersal selective disadvantage relative to individuals sired by local fathers; and (ii) the plantations’ fecundity, and consequently the proportion of non-local male gametes available for immigration into natural stands, has increased during the last years as planted trees began to reach reproductive maturity. If the first (‘migration load’) hypothesis was true, the natural selection would tend to eliminate individuals sired by non-local fathers in the long term, provided that immigration rates and selection differentials remained unchanged. If the second hypothesis was the main reason for the observed pattern, we could expect even further immigration increments in coming years, which would increase the exposure of the relict population to non-local genes and, ultimately, lead to the eventual displacement of local ones (‘gene swamping’). In order to test the migration load hypothesis, it would be necessary to carry out common garden experiments under controlled conditions, measuring the relative performance under varied environments of progenies from local mothers sired by either local or non-local pollen donors. The latter experiment represents a necessary further step, subsequent to the estimation of migration rates and their associated statistical uncertainty, to build a sounder risk assessment protocol for gene flow in conservation biology.
References
Adams WT, Hipkins VD, Burczyk J, Randall WK (1997). Pollen contamination trends in a maturing Douglas-fir seed orchard. Can J For Res 27: 131–134.
Alía R, Notivol E, Moro J (2001). Genetic variability of Spanish provenances of Scots pine (Pinus sylvestris L.): growth traits and survival. Silva Fenn 35: 27–38.
Coles S (2001). An Introduction to Statistical Modeling of Extreme Values. Springer: London.
Cornuet JM, Piry S, Luikart G, Estoup A, Solignac M (1999). New methods employing multilocus genotypes to select or exclude populations as origins of individuals. Genetics 153: 1989–2000.
Farjon A (1998). World Checklist and Bibliography of Conifers. Royal Botanic Gardens, Kew.
Faubet P, Gaggiotti OE (2008). A new bayesian method to identify the environmental factors that influence recent migration. Genetics 178: 1491–1504.
Faubet P, Waples RS, Gaggiotti OE (2007). Evaluating the performance of a multilocus Bayesian method for the estimation of migration rates. Mol Ecol 16: 1149–1166.
Hu XS, Ennos RA (1999). Impacts of seed and pollen flow on population genetic structure for plant genomes with three contrasting modes of inheritance. Genetics 152: 441–450.
Lenormand T (2002). Gene flow and the limits to natural selection. Trends Ecol Evol 17: 183–189.
Lopez S, Rousset F, Shaw FH, Shaw RG, Ronce O (2008). Migration load in plants: role of pollen and seed dispersal in heterogeneous landscapes. J Evolution Biol 21: 293–309.
Manel S, Gaggiotti OE, Waples RS (2005). Assignment methods: matching biological questions with appropriate techniques. Trends Ecol Evol 20: 136–142.
Manly BFJ (1997). Randomization, Bootstrap and Monte Carlo Methods in Biology, 2nd edn. Chapman & Hall: London.
Mercer KL, Wainwright JD (2008). Gene flow from transgenic maize to landraces in Mexico: an analysis. Agr Ecosyst Environ 123: 109–115.
Paetkau D, Calvert W, Stirling I, Strobeck C (1995). Microsatellite analysis of population structure in Canadian polar bears. Mol Ecol 4: 347–354.
Paetkau D, Slade R, Burden M, Estoup A (2004). Genetic assignment methods for the direct, real-time estimation of migration rate: a simulation-based exploration of accuracy and power. Mol Ecol 13: 55–65.
Pella J, Masuda M (2001). Bayesian methods for analysis of stock mixtures from genetic characters. Fish bull 99: 151–167.
Plomion C, LeProvost G, Pot D, Vendramin G, Gerber S, Decroocq S et al. (2001). Pollen contamination in a maritime pine polycross seed orchard and certification of improved seeds using chloroplast microsatellites. Can J Forest Res 31: 1816–1825.
Provan J, Soranzo N, Wilson NJ, McNicol JW, Forrest GI, Cottrell J et al. (1998). Gene-pool variation in Caledonian and European Scots pine (Pinus sylvestris L.) revealed by chloroplast simple-sequence repeats. P Roy Soc Lond B Bio 265: 1697–1705.
Rannala B, Mountain JL (1997). Detecting immigration by using multilocus genotypes. P Natl Acad Sci USA 94: 9197–9201.
Robledo-Arnuncio JJ, Collada C, Alía R, Gil L (2005). Genetic structure of montane isolates of Pinus sylvestris L. in a Mediterranean refugial area. J Biogeogr 32: 595–605.
Sinclair WT, Morman JD, Ennos RA (1999). The postglacial history of Scots pine (Pinus sylvestris L.) in western Europe: evidence from mitochondrial DNA variation. Mol Ecol 8: 83–88.
Slavov GT, Howe GT, Gyaourova AV, Birkes DS, Adams WT (2005). Estimating pollen flow using SSR markers and paternity exclusion: accounting for mistyping. Mol Ecol 14: 3109–3121.
Stoehr MU, Newton CR (2002). Evaluation of mating dynamics in a lodgepole pine seed orchard using chloroplast DNA markers. Can J Forest Res 32: 469–476.
Vendramin G, Lelli L, Rossi P, Morgante M (1996). A set of primers for the amplification of 20 chloroplast microsatellites in Pinaceae. Mol Ecol 5: 595–598.
Wilson GA, Rannala B (2003). Bayesian inference of recent migration rates using multilocus genotypes. Genetics 163: 1177–1191.
Acknowledgements
We thank the editor and one anonymous reviewer for constructive comments on the paper. This work was supported by Grants 59/2003 and AEG06-054 from the ‘Organismo Autónomo de Parques Nacionales’ and ‘Dirección General para la Biodiversidad’, respectively, both belonging to the Spanish ‘Ministerio de Medio Ambiente y Medio Rural y Marino’. JJRA and SCGM were supported by Ramón y Cajal research fellowships from the Spanish ‘Ministerio de Ciencia e Innovación’.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Robledo-Arnuncio, J., Navascués, M., González-Martínez, S. et al. Estimating gametic introgression rates in a risk assessment context: a case study with Scots pine relicts. Heredity 103, 385–393 (2009). https://doi.org/10.1038/hdy.2009.78
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/hdy.2009.78
Keywords
This article is cited by
-
Genetic composition of exotic and native teak (Tectona grandis) in Myanmar as revealed by cpSNP and nrSSR markers
Conservation Genetics (2016)
-
Development and transferability of two multiplexes nSSR in Scots pine (Pinus sylvestris L.)
Journal of Forestry Research (2015)
-
Space, time and complexity in plant dispersal ecology
Movement Ecology (2014)
-
Contemporary pollen-mediated gene immigration reflects the historical isolation of a rare, animal-pollinated shrub in a fragmented landscape
Heredity (2014)