Abstract
The Norfolk Island population in the South Pacific is primarily the product of recent admixture between a small number of British male and Polynesian female founders. We identified and genotyped 128 Ancestry Informative Markers (AIMs) spread across the autosomes, X/Y chromosomes and mitochondrial DNA genome, to explore and quantify the current levels of genetic admixture in the Norfolk Islanders. On the basis of autosomal AIMs, the population shows mean European and Polynesian ancestry proportions of 88 and 12%, respectively. However, there is a substantial variation between individuals ranging from total European ancestry to near total Polynesian origin. There is a strong correlation between individual genetic estimates of Polynesian ancestry and those derived from the extensive pedigree and genealogical records of Islanders. Also in line with historical accounts, there is a substantial asymmetry in the maternal and paternal origins of the Islanders with almost all Y-chromosomes of European origin whereas at least 25% of mtDNAs appear to have a Polynesian origin. Accurate knowledge of ancestry will be important in future attempts to use the Island population in admixture mapping approaches to find the genes that underlie differences in the risk to some diseases between Europeans and Polynesians.
Similar content being viewed by others
Introduction
Norfolk Island is a small volcanic island in the south Pacific located about 1600 km northeast of Sydney, Australia. The island was initially settled by Polynesians sometime between the tenth and fifteenth centuries AD, but this settlement was unsuccessful and the population later disappeared. The island was re-discovered by Captain James Cook in 1774 on his second voyage to the South Pacific and was established and settled by the British as a penal colony in 1788. It served this function for periods up until 1855 when the last convicts were relocated to Tasmania and the island abandoned (Edgecombe, 1999). In 1856, 193 residents of Pitcairn Island, also in the south Pacific, were re-settled on Norfolk Island. Pitcairn Island itself has an unusual history because the population was founded by mutineers from the legendary ‘HMS Bounty’ in 1790. The initial population consisted of Fletcher Christian, who led the mutiny, eight additional sailors, 6 men and 12 women from Tahiti, and one baby girl, for a total of 28 settlers. The Island community then developed in isolation with little immigration but by the mid nineteenth century Pitcairn was unable to support a growing population leading to the relocation of most inhabitants to Norfolk Island (Edgecombe, 1999; Hoare, 1999). The island has a strong genealogical tradition with extensive family records tracing the descent of individuals from 9 male Bounty mutineers and the 12 Tahitian females. The original Tahitian males are not thought to have left any descendents. The current population of ∼2000 individuals are thus a product of recent admixture between Europeans (primarily British) and Polynesians.
We sought to explore the relative Polynesian and European genetic ancestry of Norfolk Islanders, using a panel of novel ancestry informative markers (AIMs) spread across the entire genome and compare these estimates to those derived from genealogical records. Accurate knowledge of variation in ancestry levels will be important when using the Island population in so called ‘admixture mapping’ approaches which use differences in ancestry across the genome to find the genes that underlie diseases which have different prevalences in Europeans and Polynesians.
Materials and methods
The sample population of 600 Norfolk Islanders was assembled as described in previous studies (Bellis et al., 2008a; Bellis et al., 2008b; Bellis et al., 2005; Cox et al., 2009; Macgregor et al., 2009). Informed consent was obtained from all participants. Each individual was genotyped for 128 AIMs in four multiplexes using SequenomiPLEX Gold chemistry on a MassARRAY Compact Mass Spectrometer (Sequenom Inc., San Diego, CA, USA) essentially as previously described (Zhao et al., 2008). Individuals with >10% missing data (n=18) were discarded leaving a total sample population of 582.
The 128 AIMs were spread across the autosomes, X/Y chromosomes and mitochondrial DNA (mtDNA) genome. Autosomal (110 markers) and X-chromosome (seven markers) AIMs were selected from 3 888 075 single nucleotide polymorphisms (SNPs), genotyped in HapMap2 Release 24. The European–American [CEU] and joint East Asian (Chinese and Japanese) populations were taken as proxies for the British and Polynesian Norfolk Island parental populations, respectively. The 3000 loci that showed the greatest allele frequency differences (δ) between these populations were initially identified and from these, a set of the highest ranking δ SNPs were selected under the additional requirements: (1) No AIM be within 1 Mb of another (to ensure approximate linkage equilibrium between markers) and (2) that each chosen AIM had at least one other SNP within 50 kb (±25 Kb) in the top 3000 (to guard against database errors such as allele flipping). The average δ among AIMs was 0.78.
The Y-chromosome and mtDNA allow a specific assessment of the likely paternal and maternal ancestral origin, respectively, of each individual. Therefore, we also genotyped four Y-chromosome SNPs that define the most common British Isles haplogroups (P, encompassing R1a and R1b; I; E3b and J) (Jobling and Tyler-Smith, 2003; Karafet et al., 2008) and seven maternally inherited mtDNA SNPs that, in combination, largely define common Polynesian haplogroups (B4, B5, P1, Q and M28) (Kayser et al., 2006). These haplogroups account for 99% of the mtDNA types observed in the Polynesian population (Kayser et al., 2006). Details of AIMs and their frequency in the Norfolk Island population can be found in Supplementary Table 1.
We first examined the Norfolk Island population structure using principal component analysis (PCA) and two separate sets of comparative global populations. The first PCA was based on 51 autosomal AIMs (SNPs) that were genotyped in common between the Norfolk Islanders and the ∼1.4 million SNPs available in the 11 HapMap 3 populations (n=988). The second used 29 autosomal AIMs genotyped in common with the 51 populations (n=940) and ∼650 000 SNPs of the Human Genome Diversity Panel (HGDP) (Li et al., 2008). PCA was conducted using the EIGENSOFT package (Patterson et al., 2006; Price et al., 2006).
Individual admixture levels were estimated, for autosomal or X-chromosome SNPs separately, using the Bayesian clustering method implemented in the STRUCTURE program (Falush et al., 2003; Pritchard et al., 2000). STRUCTURE can estimate the proportion of each individual's genome derived from K number of clusters or populations using the genotype data alone. We conducted STRUCTURE runs for K=2 to K=5, using 150 000 iterations, the first 50 000 discarded as burn in. The likelihood of the data was similar for all K⩾2 and given our specific a priori hypothesis of two parental populations we focused on results for K=2.
Results and discussion
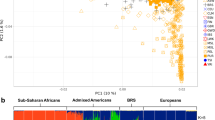
The population of Norfolk Island was initially placed in a broader genetic context by using PCA to extract the major trends/patterns present in the genotype data of islanders and individuals from other worldwide populations (Figure 1). Principal component (PC) 1 and 2 from PCA based on 51 autosomal AIMs genotyped in common between the 11 HapMap3 global populations (n=988) and the Norfolk Islanders is shown in Figure 1a. As these SNPs were selected based on their ability to differentiate Europeans and East Asians, it is unsurprising that the major trend (PC 1) separates these groups from each other. Although most Norfolk Islanders fall among the two European population samples (European–Americans (CEU) and Tuscan–Italians (TSI)), others range along PC1 with a small number grouping closely with the East Asian populations (Chinese from Beijing (CHB) and Denver (CHD) and the Japanese (JPT)). These observations are consistent with predominant European ancestry but also varying levels of East Asian input between individuals. The latter presumably reflects Polynesian ancestry because there is no evidence of more recent direct East Asian migration after the foundation of the Norfolk Island population. A similar result is seen along PC1 in PCA of populations from the HGDP (Figure 1b). However, there is a somewhat poorer resolution of major population clusters, compared with HapMap3, because analysis is based on fewer (29) SNPs that were genotyped in both datasets.
(a) Principal component analysis (PCA) of Norfolk Islanders (n=582) and unrelated individuals from the 11 global HapMap3 (n=988) populations using 51 autosomal SNPs. HapMap3 population codes and samples sizes are as follows: ASW-African ancestry from Southwest USA, n=49; CEU-Utah residents with Northern and Western European ancestry from the CEPH collection, n=112; CHB-Han Chinese in Beijing, China, n=84; CHD-Chinese in Metropolitan Denver, Colorado, n=85; GIH-Gujarati Indians in Houston; Texas, n=88; JPT-Japanese in Tokyo, Japan, n=86; LWK-Luhya in Webuye, Kenya, n=90; MEX-Mexican ancestry in Los Angeles; California, n=50; MKK-Maasai in Kinyawa, Kenya, n=143; TSI-Tuscans, Italy, n=88; YRI-Yoruba in Ibadan, Nigeria n=113. (b) PCA of Norfolk Islanders and individuals from the HGDP (n=940) populations using 29 autosomal SNPs genotyped in common between both datasets. The 51 populations of the HGDP have been divided into seven regional groupings (see Li et al., 2008). Their codes and samples sizes are as follows: AFR-Africa, n=101; AME-America, n=64; CSA-Central-Southern Asia, n=200; ESA-Eastern Asia, n=228; EUR-Europe, n=157; MDE-Middle-East, n=163; OCE-Oceania (Melanesians and Papuans), n=27.
Quantification of individual autosomal genomic admixture was undertaken using STRUCTURE and 110 autosomal AIMs (the remaining 18 AIMs were spread across the X-chromosome, Y-chromosome and mtDNA genome) under a model of two parental populations (K=2). We had first investigated the discriminatory power of the AIM set by carrying out STRUCTURE analysis using only the 51 AIMs genotyped in common with the Tuscan–Italian (TSI) and Chinese–Denver (CHD) population samples from HapMap3 which were not used in the initial ascertainment of the AIMs. The individuals from the two populations were correctly assigned to different clusters with an average individual ancestry of >0.995.
The HapMap2 European–American (CEU) sample is of North/West Europe ancestry and is a good proxy for the European parental population of Norfolk Island because the Bounty mutineers were from the British Isles. The use of the East Asian populations as a proxy for the Polynesian parental population is less obviously relevant. However, genetic evidence suggests that Polynesians are predominantly (70–80%) East Asian in origin, stemming from the expansion of Austronesian speakers across the region beginning ∼3000 years ago (Kayser et al., 2008; Kimura et al., 2008). As the process of island settlement across the south Pacific, and beyond, likely involved repeated ‘bottleneck’ founder effects (where only a subset of original migrants made each subsequent step), allele frequencies may have changed because of random sampling effects (genetic drift) compared to those observed in the current East Asian population. The remaining 20–30% of Polynesian ancestry results from inter-breeding with the earlier indigenous people of island Oceania (Melanesians). Although Melanesian populations have a distinct history they are expected to show greater co-ancestry with East Asians than Europeans. We observe this along PC1 in the HGDP populations (which includes East Asians, Melanesians and Europeans) using a subset of SNPs from our AIM set (Figure 1b). Although the ascertainment of AIMs using the East Asian population could reduce power to detect Polynesian ancestry we conclude that the effect is likely to be minimal and somewhat offset by the large pool of SNPs (∼4 million) available in HapMap2, which aids the initial identification of very informative AIMs (those with the greatest allele frequency differences between parental populations).
The distribution of Norfolk Islander STRUCTURE cluster 1 admixture proportions, which as the majority component (average of 88%) represents European ancestry, is shown in Figure 2a. There is a strong correlation between PC1 scores, derived from PCA with the HapMap3 populations, and STRUCTURE European ancestry fractions for each individual (r=0.91). Consistent with the PCA, about half of the Island inhabitants (51%) have little/negligible (<5%) Polynesian ancestry (and are conversely >95% European), with fewer individuals displaying a range of admixture proportions up to 94% Polynesian. On the basis of genealogical information, the sample may be subdivided into two major groupings. Cohort 1 (herein termed ‘Pedigree’, n=324), are connected together in a single large pedigree from 11 founding males (9 ‘Bounty’ mutineers and two Europeans who arrived in the mid nineteenth century) and six Tahitian women (Macgregor et al., 2009). The other major cohort (termed ‘non-pedigree’, n=258) are more recent, mainly twentieth century and largely unrelated, migrants to the Island, who were predominantly of European ancestry (generally from Australian and New Zealand). Consistent with the known history, the distribution of individual European ancestry estimates between the two groups is very different (Figure 2a) (P ⋜1.6 × 10−24, two-sample t-test). For instance 81% of non-pedigree individuals have little reliably detectable (<5%) Polynesian ancestry whereas the equivalent figure for pedigree individuals is just 27%.
(a) Distribution of individual European ancestry estimates for the entire sample (‘All’, n=582), those which are part of a large pedigree from the original founders (‘Pedigree’, n=324) and more recent twentieth century migrants (‘Non-Pedigree’, n=258). Corresponding Polynesian ancestry estimates are (1-European component). (b) Overall European ancestry estimates from different genomic markers. Those from the autosomes and X-chromosome are average cluster 1 (European) values from STRUCTURE analysis. Results for the Y-chromosomes are the fraction of all fully genotyped male individuals (n=250) whose Y-chromosome falls into one of the four typical British Isles/Western European haplogroups (P, encompassing R1a and R1b; I; E3b and J) and as such represents the minimum European contribution. mtDNA results are the fraction of individuals fully genotyped for a suite of seven mtDNA SNPs (n=573) who do not show the motifs for typical Polynesian haplogroups (B4, B5, P1, Q and M28). As this assumes that all other mtDNA originated in Europe, the illustrated values are conservative and represent the maximum European contribution.
In line with this, the overall average European autosomal admixture proportions are greater in the non-pedigree versus pedigree individuals, 95 versus 82%, respectively, with an overall average of 88% (Figure 2b). Conversely, Polynesian ancestry is estimated at 5 and 18% in the non-pedigree and pedigree cohorts, respectively (average 12% in the entire sample). The autosomal genome is a composite of all ancestors of an individual. However, as the X-chromosome, Y-chromosome and mtDNA have different inheritance patterns, they provide additional insights into gender asymmetry in the population history of Norfolk Island. The Y-chromosome is paternally inherited and as expected from history, the Y-chromosomes of Norfolk Islanders are consistent with all European male founders, considering only four major European haplogroups were tested (and therefore we can only detect the minimum European Y-chromosome ancestry, Figure 2b). Maternally inherited mtDNA suggests that a minimum of 23% of the total population samples have a direct Polynesian maternal ancestor, which encompasses ∼40% of those in the main pedigree and just ∼4% of non-pedigree Islanders. Assuming an equal sex ratio, ∼25% female and 0% male Polynesian founders are expected to yield the ∼12% Polynesian autosomal ancestry estimated by STRUCTURE. Each X-chromosome spends 2/3 of its history in females and therefore might be expected to show admixture proportions intermediate between autosomal and mtDNA. Although X-chromosomes did show slightly, but consistently, lower European ancestry, these STRUCTURE estimates are based on just seven markers and thus may lack the depth of precision required to differentiate to such a level (Figure 2b).
A previous study estimated the level of Polynesian admixture for each individual in the main founder pedigree based on the genealogical structure of the pedigree itself. Estimates of Polynesian ancestry ranged from 0–45% (Macgregor et al., 2009). We compared these estimates to those derived from the set of autosomal AIMs (Figure 3). There is a highly significant correlation between the two sets of ancestry estimates (r=0.72; P<0.05) although there is clearly some discrepancy for some individuals. Minor discrepancies are likely to result from inherent uncertainty in the precision of both genetic and pedigree-based ancestry estimates whereas major discrepancies could result from errors in the genealogy or the assumption in the pedigree estimates that later married-in individuals were all European.
Relationship between pedigree and genetic-based estimates of Polynesian Ancestry (r=0.72, P<0.05). Pedigree estimates were made in a previous study and are the fraction of each Islander's genome derived from six founding Polynesian females according to genealogical records (Macgregor et al., 2009). Genetic estimates are the proportion of each Islander's genome assigned to Cluster 2, presumed Polynesian ancestry, based on STRUCTURE analysis of 110 autosomal AIMs.
The results genetically characterize the extent and nature of European and Polynesian admixture during the short history of the Norfolk Island population. Genetic ancestry estimates concur remarkably well with both the overall island population history and genealogical records for specific individuals. Variation in genomic ancestry levels are the basis of ‘admixture mapping’ approaches, which may be appropriate in the Norfolk Island population to find the genes involved in phenotypes, for example various cardiovascular disease indicators, that differ in frequency between the European and Polynesian parental populations.
References
Bellis C, Cox HC, Dyer TD, Charlesworth JC, Begley KN, Quinlan S et al. (2008a). Linkage mapping of CVD risk traits in the isolated Norfolk Island population. Hum Genet 124: 543–552.
Bellis C, Cox HC, Ovcaric M, Begley KN, Lea RA, Quinlan S et al. (2008b). Linkage disequilibrium analysis in the genetically isolated Norfolk Island population. Heredity 100: 366–373.
Bellis C, Hughes RM, Begley KN, Quinlan S, Lea RA, Heath SC et al. (2005). Phenotypical characterisation of the isolated norfolk island population focusing on epidemiological indicators of cardiovascular disease. Hum Hered 60: 211–219.
Cox HC, Bellis C, Lea RA, Quinlan S, Hughes R, Dyer T et al. (2009). Principal component and linkage analysis of cardiovascular risk traits in the Norfolk isolate. Hum Hered 68: 55–64.
Edgecombe J (1999). Norfolk Island—South Pacific : Island of History and Many Delights. Australia: Thornleigh.
Falush D, Stephens M, Pritchard JK (2003). Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics 164: 1567–1587.
Hoare M (1999). Norfolk Island: A Revised and Enlarged History 1774–1998. Central Queensland University Press: Rockhampton.
Jobling MA, Tyler-Smith C (2003). The human Y chromosome: an evolutionary marker comes of age. Nat Rev Genet 4: 598–612.
Karafet TM, Mendez FL, Meilerman MB, Underhill PA, Zegura SL, Hammer MF (2008). New binary polymorphisms reshape and increase resolution of the human Y chromosomal haplogroup tree. Genome Res 18: 830–838.
Kayser M, Brauer S, Cordaux R, Casto A, Lao O, Zhivotovsky LA et al. (2006). Melanesian and Asian origins of Polynesians: mtDNA and Y chromosome gradients across the Pacific. Mol Biol Evol 23: 2234–2244.
Kayser M, Lao O, Saar K, Brauer S, Wang X, Nurnberg P et al. (2008). Genome-wide analysis indicates more Asian than Melanesian ancestry of Polynesians. Am J Hum Genet 82: 194–198.
Kimura R, Ohashi J, Matsumura Y, Nakazawa M, Inaoka T, Ohtsuka R et al. (2008). Gene flow and natural selection in oceanic human populations inferred from genome-wide SNP typing. Mol Biol Evol 25: 1750–1761.
Li JZ, Absher DM, Tang H, Southwick AM, Casto AM, Ramachandran S et al. (2008). Worldwide human relationships inferred from genome-wide patterns of variation. Science 319: 1100–1104.
Macgregor S, Bellis C, Lea RA, Cox H, Dyer T, Blangero J et al. (2009). Legacy of mutiny on the bounty: founder effect and admixture on Norfolk Island. Eur J Hum Genet (in press; e-pub ahead of print 8 July 2009; doi:10.1038/ejhg.2009.111).
Patterson N, Price AL, Reich D (2006). Population structure and eigenanalysis. PLoS Genet 2: e190.
Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D (2006). Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet 38: 904–909.
Pritchard JK, Stephens M, Donnelly P (2000). Inference of population structure using multilocus genotype data. Genetics 155: 945–959.
Zhao ZZ, Nyholt DR, Thomas S, Treloar SA, Montgomery GW (2008). Polymorphisms in the vascular endothelial growth factor gene and the risk of familial endometriosis. Mol Hum Reprod 14: 531–538.
Acknowledgements
We thank Norfolk Island volunteers for their participation in this study and staff in the QIMR Molecular Epidemiology laboratory for preparation of samples for genotyping. This work was supported by grants from the Australian National Health and Medical Research Council (Medical Genomics Program Grant 389892) and the Griffith Medical Research College (a joint program of Griffith University and the Queensland Institute of Medical Research, QIMR, Herston, QLD, Australia, 4006). SM, GWM and PMV are supported by the NHMRC fellowship scheme.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies the paper on Heredity website
Supplementary information
Rights and permissions
About this article
Cite this article
McEvoy, B., Zhao, Z., Macgregor, S. et al. European and Polynesian admixture in the Norfolk Island population. Heredity 105, 229–234 (2010). https://doi.org/10.1038/hdy.2009.175
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/hdy.2009.175
Keywords
This article is cited by
-
Pedigree derived mutation rate across the entire mitochondrial genome of the Norfolk Island population
Scientific Reports (2022)
-
‘Mutiny on the Bounty’: the genetic history of Norfolk Island reveals extreme gender-biased admixture
Investigative Genetics (2015)
-
Serum bilirubin concentration is modified by UGT1A1 Haplotypes and influences risk of Type-2 diabetes in the Norfolk Island genetic isolate
BMC Genetics (2015)