Abstract
Triploid loaches Misgurnus anguillicaudatus are derived from unreduced diploid gametes produced by an asexual clonal lineage that normally undergoes gynogenetic reproduction. Here, we have investigated the reproductive system of two types of triploids: the first type carried maternally inherited clonal diploid genomes and a paternally inherited haploid genome from the same population; the second type had the same clonal diploid genomes but a haploid genome from another, genetically divergent population. The germinal vesicles of oocytes from triploid females (3n=75) contained only 25 bivalents, that is, 50 chromosomes. Flow cytometry revealed that the majority of the progeny resulting from fertilization of eggs from triploid females with normal haploid sperm were diploid. This indicates that triploid females mainly produced haploid eggs. Microsatellite analyses of the diploid progeny of triploid females showed that one allele of the clonal genotype was not transmitted to haploid eggs. Moreover, the identity of the eliminated allele differed between the two types of triploids. Our results demonstrate that there is preferential pairing of homologous chromosomes as well as the elimination of unmatched chromosomes in the course of haploid egg formation, that is, meiotic hybridogenesis. Two distinct genomes in the clone suggest its hybrid origin.
Similar content being viewed by others
Introduction
Interspecific hybridization often causes a disruption of normal meiosis and resultant sterility due to the failure of pairing between homologous chromosomes. In lower vertebrates, some hybrids have escaped from this reproductive constraint by acquiring an atypical mode of reproduction such as unreduced gametogenesis and parthenogenesis or gynogenesis (Dawley, 1989; Vrijenhoek et al., 1989; Beukeboom and Vrijenhoek, 1998). The loach, Misgurnus anguillicaudatus (Teleostei:Cobitidae), reproduces both sexually and asexually. In the latter case, diploid females (2n=50) spawn genetically identical, unreduced diploid eggs, most of which develop gynogenetically. However, some eggs produce triploid (3n=75) progeny following the accidental incorporation of a haploid sperm nucleus after fertilization (Morishima et al., 2002; Itono et al., 2006, 2007). The relationship between atypical reproduction and hybridization is uncertain in the loach because Japanese M. anguillicaudatus has been taxonomically identified as a single species entity (Saitoh, 1989). Thus it is very difficult to explain the occurrence of a natural, asexually reproducing clone by the hybrid-origin theory. However, analyses of the sequences of the mtDNA control region clearly indicated the presence of two different clades, probably corresponding to two species in the phylogenic tree. This strongly suggests that Japanese M. anguillicaudatus might be comprised of at least two cryptic species (Morishima et al., 2008a). Currently, the evolutionary status of the clonal loach remains uncertain. The genomic or specific constitution has been estimated in unisexual and polyploid biotypes of fish species, belonging to the genera Poeciliopsis, Poecilia, Menidia, Phoxinus and Cobitis (Vrijenhoek et al., 1989). In contrast, the Iberian minnow Squalius (Rutilus) alburnoides has been clearly shown to be a complex that originated by intergenetic hybridization between an Squalius pyrenaicus female and a male of a probably extinct species similar to Anacypris hispanica (Alves et al., 2001, 2002, 2004).
Allotriploid hybrid animals have two genomes (chromosome sets) of one species and one genome of a second species. During meiosis, the chromosomes of the former species often show preferential pairing (formation of bivalents) because they share greater homology, while the chromosomes of the second species remain unpaired. The bivalents segregate to produce haploid gametes, whereas unpaired chromosomes may be excluded. This type of gametogenetic system was first proved in the Iberian minnow by using molecular markers and defined as meiotic hybridogenesis by Alves et al. (1998), although the possible involvement of this system was formerly suggested in the natural triploid hybrid frog Rana esculenta (Günther et al., 1979) and subsequently in artificial triploid hybrid frogs (Nishioka and Ohtani, 1984; Ohtani, 1993). A similar pattern of reproduction has also been observed in other triploid hybrid fishes of the genus Phoxinus (Goddard and Schultz, 1993) and Cobitis (Kim and Lee, 2000; Saitoh et al., 2004). Meiotic hybridogenesis differs from “normal” hybridogenesis that is typically found in hemi-clonally reproducing hybrid fish of the genus Poeciliopsis (Schultz, 1961, 1966, 1969), in which an intact maternal haploid genome is transmitted to the eggs and the paternal haploid genome is eliminated (Cimino, 1972).
Although induced triploid fish are generally sterile (Arai, 2001), a triploid female derived from an unreduced diploid egg of the clonal loach after normal fertilization was reported to spawn haploid eggs that developed to normal diploid progeny after fertilization with haploid sperm (Oshima et al., 2005). If a clone contains two different genomes inherited from two different cryptic species, as suggested by mtDNA sequences (Morishima et al., 2008a), sperm incorporation by diploid eggs of the clonal females may generate allotriploids.
In the present study, we prepared two types of triploid loach with different genomic constitutions; both types had in common the diploid genomes from the clonal lineage, but had a different haploid genome. We confirmed the production of haploid eggs by cytological observation of the meiotic chromosomes in the germinal vesicle of full-grown oocytes from the two types of triploid females. In addition, we carried out a genotypic analysis of 10 microsatellite loci (nine of which mapped to independent linkage groups, that is, different chromosomes; Morishima et al., 2008b) in the diploid progeny of the triploid females to examine the segregation pattern of alleles inherited from the natural clone.
Materials and methods
Fish samples
The triploid females 3NF1 and 3NF2 were collected in 2005 from a wild population in Ozora (previously called Memanbetsu), in the northern area of Hokkaido Island, Japan. Flow cytometric analysis of DNA contents was used to determine ploidy status as described previously (Morishima et al., 2002). In this population, the results of mtDNA analysis indicate that normal bisexual diploids predominantly belong to clade A (Morishima et al., 2008a). The triploid females 3NF3 and 3NF4 were generated by fertilization using unreduced diploid sperm from a sex-reversed diploid clonal loach (Yoshikawa et al., 2007) and the normal haploid eggs of bisexual diploids taken from a paddy field in Hokuto (previously called Ohno) in the southern area of Hokkaido Island in 2003. Mitochondrial DNA sequence analysis indicates that bisexual diploids in the southern area of Hokkaido Island belong to clade B (Morishima et al., 2008a). Thus, triploid 3NF1 and 3NF2 may have two genomes transmitted from the clone and one genome of a bisexual loach of clade A, whereas triploid 3NF3 and 3NF4 have two genomes inherited from the clone and one genome of a clade B loach. We named 3NF1 and 3NF2 as type I triploids and 3NF3 and 3NF4 as type II triploids.
Mature eggs laid by triploid females 3NF1, 3NF2 and 3NF3 were fertilized by haploid sperm to produce the progeny in families no. 1, no. 2 and no. 3, respectively. Triploid female 3NF4 did not spawn mature eggs at this trial. Haploid sperm was collected from normal bisexual diploid loach cultured from fish originally taken in the southern part of Hokkaido. Artificial ovulation and fertilization followed the standard procedure described by Morishima et al. (2002). The ploidy status of selected progeny of the triploid females (about 10 individuals per family) was determined by DNA-content flow cytometry as described previously (Morishima et al., 2002). The ovaries of 3NF1, 3NF2 and 3NF4 were used for cytological observation of meiotic chromosomes (see the following section). The ovaries of two diploid fish, 2NF1 and 2NF2, from Hokuto were used as a control.
Cytological observation of meiotic chromosomes
Cytological observation was performed according to the methods previously described by Itono et al. (2006). The females were injected with 20–25 IU hCG (Teikoku Hormone Medical Co. Ltd., Kawasaki, Japan) per gram body weight and kept for 4–5 h at 25 °C in an aquarium. Full-grown oocytes were isolated from the ovary and then incubated in physiological saline for goldfish containing 17α–20β dihydroxy-4-pregnene-3-one (Sigma-Aldrich, St Louis, MO, USA) at room temperature. During the period from germinal vesicle migration to immediately prior to germinal vesicle breakdown, the oocytes were periodically fixed with Carnoy's fixative and the germinal vesicle (GV) was dissected out under a stereoscopic microscope. The isolated GV was placed on a clean glass slide, air dried and then stained with DAPI (4′,6-diamidino-2-phenylindole) (Wako, Osaka, Japan) for 1 h. Meiotic chromosomes were viewed using a fluorescence microscope.
Microsatellite genotyping
Progeny of triploid females 3NF1, 3NF2 and 3NF3 were used for microsatellite genotyping. Ten diagnostic microsatellite DNA, of the 97 microsatellite markers developed and mapped to linkage groups (Morishima et al., 2001, 2008b), were screened to distinguish all three alleles of each triploid female. Eight of the markers identify loci on independent linkage groups, that is, different chromosomes; Mac73 and Mac204 both map to linkage group 9 (Morishima et al., 2008b).
M13M1 plus GCC primers that were fluorescently labeled with NED, VIC, FAM or PET (Applied Biosystems, Foster, CA, USA) were used for genotyping. All of the forward primers had an M13M1 tail sequence (5′-AGTCACGACGTTGTA-3′) attached at their 5′ ends. PCR was carried out in a reaction mixture (10 μl) containing 25 ng template DNA, 0.025 U of Taq DNA polymerase (TaKaRa), 40 μM dNTPs, 0.3 pmol M13M1-tailed forward primer, 3.0 pmol reverse primer and 3.0 pmol M13M1 primer. The following conditions were used: 1 cycle of initial denaturation for 1 min at 93 °C 35–40 cycles of denaturation for 30 s at 93 °C, annealing for 30 s at 56 °C, extension for 30 s at 72 °C and 1 cycle of final extension for 60 min at 72 °C. The PCR products were stored at 4 °C until the genotyping process. Electrophoresis was carried out using the automated sequencer ABI3130xl (Applied Biosystem). The alleles were distinguished by differences in molecular size (bp: base pair) using the Gene Scan 500 LIZ size standard (Applied Biosystem). Genotyping was performed using the GeneMapper program Ver.3.7 (Applied Biosystem).
Estimation of recombination rate
Recombination rates between the Mac73 and Mac204 loci on linkage group (LG) 9 (Morishima et al., 2008b) were calculated by using the program Mapmanager QTLXb20 (http://:www.mapmanager.org/mmQTX.htm). In this analysis, the pseudo-cross test (Grattapaglia and Sederoff, 1994) was used because the phase or the origin of gametes was unknown.
Results
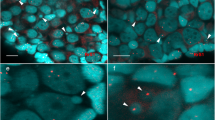
The majority of germinal vesicles (GVs) isolated from the ovaries of two diploid females (2n=50), 2NF1 and 2NF2 and the three triploid females (3n=75), 3NF1, 3NF2 and 3NF4, contained 25 bivalents (Table 1, Figures 1a and b). Thus, 50 of the 75 chromosomes in triploids paired to form bivalents, as in the control diploids; however, the 25 unpaired chromosomes expected to be present in addition to the bivalents were not found. A few metaphases from the GVs of triploid females contained approximately 75 bivalents (Table 1, Figure 1c), but trivalents, univalents and other unusual meiotic configurations were not detected.
Paired chromosomes (bivalents) in germinal vesicles (GVs) isolated from oocytes of ovaries of control and triploid female. (a) GV of a diploid control showing 25 bivalents; (b) GV of triploid female showing 25 bivalents with no indication of the additional genome; (c) GV of triploid female showing approximately 75 bivalents (73 bivalents countable). Scale bar, 20 μm.
All of the progeny (n=10) from each family were determined by flow cytometry to be diploid and most had diploid genotypes at the tested microsatellite loci. As is shown in Table 2, the three different alleles at each microsatellite locus examined in each triploid female were not transmitted at random to the haploid eggs, and the pattern of preferential allelic segregation was different between the type I (3NF1 and 3NF2) and type II (3NF3) triploids. At the Mac3 locus, for example, 3NF1 and 3NF3 had tri-allelic genotypes (103/145/125 and 103/145/105, respectively); thus, both included the identical clonal diploid genotypes 103/145 (Table 2). In the diploid progeny of 3NF1 (family no. 1, type I), alleles 103 and 125, but not 145, were observed (Table 2). In contrast, in the progeny of 3NF3 (family no. 3, type II), alleles 145 and 105 appeared, but allele 103 was not transmitted (Table 2). Thus, allele 145 was not transmitted by type I triploid females, whereas allele 103 was not transmitted by type II triploid females. For all 10 loci examined (located on nine separate linkage groups; Morishima et al., 2008b), a similar pattern of preferential segregation of alleles was observed.
Thus, in the type I triploid females, one genome derived from the clone (and tentatively named genome A′) paired with genome A from the loach of the same population. In contrast, the second genome derived from the clone (and tentatively named B′) was eliminated (Table 2). The genomic constitution of type I triploid females is thus described as A′B′A. Similarly, the genomic constitution of type II triploid females is described as A′B′B as the microsatellite allele from genome B of the genetically different loach was transmitted to the gametes, but the allele from genome A′ in the clone was eliminated (Table 2). However, this selective segregation was not perfect and a few exceptions were recovered, such as two examples with 107/113 alleles at Mac37 in the progeny of the 3NF2 female (Table 2).
A small number of triploid or trisomic aneuploid progeny were detected by the presence of a tri-allelic genotype at one or more loci examined (Table 2). A total of two, six and three triploid or aneuploid progeny were identified in families numbers 1, 2 and 3, respectively. Five of the non-diploid progeny from family no. 2 had tri-allelic genotypes at four to six loci, the other at only one locus. The two non-diploid progeny from family no. 1, one from no. 2 and three from no. 3 exhibited tri-allelic genotypes at only one or two loci. Since the triploid or trisomic aneuploid progeny carried two alleles of maternal origin and one allele of paternal origin, we could discount the possibility that they were gynogenotes. In eight of these 11 non-diploid progeny, non-clonal genotypes (ac or bc in Table 2) were transmitted to mature eggs. The clonal genotype (ab) was seen in only three progeny (Table 2).
The recombination rate between Mac73 and Mac204 on LG9 (Morishima et al., 2008b) was estimated to be 0 cM in two males for family no. 1 and no. 2, 5.1 cM in 3NF1 female and 2.5 cM in 3NF2 female. The recombination rate in family no. 3 was not estimated because three alleles at each locus could not be distinguished. Four recombinants were identified among the 78 progeny of 3NF1 (family no. 1), whereas two recombinants were present among 80 progeny examined for both loci in 3NF2 (family no. 2). Thus, recombination can occur between a chromosome from the clone and another derived from an accidentally incorporated genome (Table 3).
Discussion
Oshima et al. (2005) examined the reproductive capacity of two triploid females captured from a natural population and reported that one laid a large number of haploid eggs and a few diploid eggs, but the other produced many aneuploid and few triploid eggs. In the present study, however, the triploid loaches analyzed predominantly produced haploid eggs. A predisposition to production of haploid eggs by triploid loach was previously reported for fish derived by hybridization between normal diploid females and natural tetraploid males (Arai and Mukaino, 1997).
The present cytological analysis demonstrated the presence of only 25 bivalents in the majority of germinal vesicles of oocytes from triploid females. This suggests that chromosome pairing in meiosis occurred between chromosomes from two of the three genomes of the triploid, and that the third genome was eliminated. Preferential pairing between two matched sets of homologous chromosomes presumably occurred during meiosis in the triploid as in the bisexual diploid; the distinguishing characteristic of the former is the elimination of one genome. As mentioned earlier, a similar meiotic hybridogenesis system has been reported previously in allotriploid vertebrates. Since no univalents were detected in the germinal vesicles of the triploid females examined, one genome may have been eliminated in the stages before the resumption of the first meiotic division in full-grown oocytes.
The microsatellite analysis identified a non-random or selective pattern of allelic segregation in triploid females. One of the three alleles had disappeared in the progeny at all loci examined. The microsatellite loci used in the present study map to nine of the presumptive 25 linkage groups in the loach (Morishima et al., 2008b). Thus, they covered about one-third of the genome, and the behavior of alleles at each locus might reflect that of each of the different chromosomes. One of the two alleles derived from the natural clone was not inherited by mature haploid eggs of triploids at any of the 10 loci examined. These results are consistent with the suggestion that only two of the genomes are involved in meiotic pairing and that one genome, derived from the clone, is eliminated during oogenesis in triploid females. The genetic results using molecular markers strongly support preferential pairing between homologous chromosomes with greater homology and the subsequent occurrence of meiotic hybridogenesis in the triploid loach. The two types of triploids differed with respect to the genome that was preferentially eliminated. These results indicate that the degree of homology between the maternally- and paternally-derived chromosome sets is the determining factor that decides which genome is eliminated.
The alleles eliminated and segregated were different in types I and II triploids (Table 2). A difference in the segregated alleles of the clone in the two types of triploids was observed at all loci: one genome (A′) of the clone is more likely to pair to the genome from the clade A loach, whereas the B′ genome is matched to the genome from clade B loach. This suggests that the genomic or chromosomal constitution of the natural clone may be of hybrid origin. We suggest that the A′ and B′ genomes show preferential pairing with the genome from the diploid loach and, as a result, the unpaired A′ or B′ genome is discarded. Some support for the suggestion that closely related genomes, A and A′ or B and B′, pair preferentially comes from the fact that A′B′ genotypes were rare in non-diploid (triploid or trisomic aneuploid) progenies of triploid females. Thus, the microsatellite alleles of the clone could be assigned to two different origins. These nuclear markers may be useful for further population studies to identify the exact origin of the clonal loach.
Our cytogenetic and genetic results indicate that the chromosomes of the A′ and B′ genomes may have a reduced affinity to pair at meiosis. This presumptive meiotic behavior by chromosomes of a natural clone cannot be explained as a consequence of karyological differentiation between populations of the loach, because the diploid karyotype of the clonal loach has been reported to be morphologically identical to that of normal bisexual loaches taken from different populations (Itono et al., 2006). The mechanisms of how homologous chromosomes recognize their counterparts remain to be elucidated.
One chromosome set is discarded prior to the resumption of meiosis in the triploid loach, but the exact timing of this elimination event was not determined in the present study. Checkpoint mechanisms that monitor irregular chromosome constitutions in triploid germ cells might alter the meiotic process to trigger elimination of an unmatched genome for the formation of regular bivalents between only matched chromosomes. In the diploid clonal loach (2n=50), premeiotic endomitosis can result in chromosome doubling without cytokinesis. As a result, these oocytes form 50 bivalents, twice the number usually observed in normal bisexual control diploids, and produce diploid eggs (Itono et al., 2006). In this system, the checkpoint is likely to trigger premeiotic endomitosis to form bivalents between doubled sister chromosomes. A small number of germinal vesicles with approximately 75 bivalents were observed in the triploid females. This suggests that premeiotic endomitosis has occurred and may result in the possible formation of rare triploid eggs. To date, however, no tetraploid progeny that might have arisen from the fertilization of triploid eggs with haploid sperm have been identified in any family. Unreduced triploid eggs have been reported in previous studies on the reproductive capacity of the natural triploid loach (Zhang et al., 1998; Zhang and Arai, 1999; Oshima et al., 2005) and triploid Iberian minnow (Alves et al., 2004).
In apomictic plants such as dandelions Taraxacum officinale, the gene controlling asexual reproduction has been predicted (Tas and van Dijk, 1999; van Dijk et al., 1999) and the sex-specific dominant gene DIPROSPOROUS was recently reported to control the formation of unreduced spores (van Dijk and Bakx-Schotman, 2004). In asexual vertebrates, however, the incidence of atypical reproduction may be controlled by irregular genomic constitution due to hybridization and polyploidy rather than the expression of a mutant gene to control the modification of meiotic events. Although various regulatory molecules necessary for normal meiosis have been actively identified in vertebrates including mammal (Eijpe et al., 2003; Baudat and de Massy, 2007) and fish (Iwai et al., 2004, 2006), little is known about probable presence of checkpoints for irregular chromosome constitutions at early germ cells that might trigger a switch in the normal meiotic process to an atypical reproductive mode, such as unreduced gametogenesis or meiotic hybridogenesis, in lower vertebrates. Natural clonal loach and its derivative polyploids are good model animals to investigate cellular and molecular mechanisms for modified gametogenesis and atypical reproduction.
References
Alves MJ, Coelho MM, Collares-Pereira MJ (1998). Diversity in the reproductive modes of females of the Rutilus alburnoides complex (Teleostei, Cyprinidae): a way to avoid the genetic constraints of uniparentalism. Mol Biol Evol 15: 1233–1242.
Alves MJ, Coelho MM, Collares-Pereira MJ (2001). Evolution in action through hybridization and polyploidy in an Iberian freshwater fish: a genetic review. Genetica 111: 375–385.
Alves MJ, Collares-Pereira MJ, Dowling TE, Coelho MM (2002). The genetics of maintenance of an all-male lineage in the Squalius alburnoides complex. J Fish Biol 60: 649–662.
Alves MJ, Gromicho M, Collares-Pereira MJ, Crespo-Lopez E, Coelho MM (2004). Simultaneous production of triploid and haploid eggs by triploid Squalius alburnoides (Teleostei: Cyprinidae). J Exp Zool 301A: 552–558.
Arai K (2001). Genetic improvement of aquaculture finfish spieces by chromosome manipulation techniques in Japan. Aquaculture 197: 205–228.
Arai K, Mukaino M (1997). Clonal nature of gynogenetically induced progeny of triploid (diploid x tetraploid) loach Misgurnus anguillicaudatus (Pisces: Cobitididae). J Exp Zool 278: 412–421.
Baudat F, de Massy B (2007). Regulating double-stranded DNA break repair towards crossover or non-crossover during mammalian meiosis. Chromosome Res 15: 565–577.
Beukeboom LW, Vrijenhoek RC (1998). Evolutionary genetics and ecology of sperm-dependent parthenogenesis. J Evol Biol 11: 755–782.
Cimino MC (1972). Egg production, polyploidization, and evolution in a diploid all-ffemale fish of the genus Poeciliopsis. Evolution 26: 294–306.
Dawley RM (1989). An introduction to unisexual vertebrates. In: Dawley RM, Bogart T JP (eds). Evolution and Ecology of Unisexual Vertebrates. New York State Museum: Albany. Bulletin 466, pp 1–18.
Eijpe M, Offenberg H, Jessberger R, Revenkova E, Heyting C (2003). Meiotic cohesin REC8 marks the axial elements of rat synaptonemal complexes before cohesins SMC1beta and SMC3. J Cell Biol 160: 657–670.
Goddard KA, Schultz RJ (1993). Aclonal reproduction by polyploid members of the clonal hybrid species Phoxinus eos-neogaeus. (Pisces; Cyprinidae). Copeia 1993: 650–660.
Grattapaglia D, Sederoff R (1994). Genetic linkage maps of Eucalyptus grandis and Eucalyptus urophylla using a pseudo-testcross: mapping strategy and RAPD markers. Genetics 137: 1121–1137.
Günther R, Uzzell T, Berger L (1979). Inheritance patterns in triploid Rana ‘esculenta’ (Amphibia, Salientia). Mitt Zool Mus Berl 55: 35–57.
Itono M, Morishima K, Fujimoto T, Bando E, Yamaha E, Arai K (2006). Premeiotic endomitosis produces diploid eggs in the natural clone loach, Misgurnus anguillicaudatus (Teleostei: Cobitidae). J Exp Zool 305A: 513–523.
Itono M, Okabayashi N, Morishima K, Fujimoto T, Yoshikawa H, Yamaha E et al. (2007). Cytological mechanisms of gynogenesis and sperm incorporation in unreduced diploid eggs of the clonal loach, Misgurnus anguillicaudatus (Teleostei: Cobitidae). J Exp Zool 307A: 35–50.
Iwai T, Lee J, Yoshii A, Yokota T, Mita K, Yamashita M (2004). Changes in the expression and localization of cohesin subunits during meiosis in a non-mammalian vertebrate, the medaka fish. Gene Express Patterns 4: 495–504.
Iwai T, Yoshii A, Yokota T, Sakai C, Hori H, Kanamori A et al. (2006). Structural components of the synaptonemal complex, SYCP1, and SYCP3, in the medaka fish Oryzias latipes. Exp Cell Res 312: 2528–2537.
Kim IS, Lee EH (2000). Hybridization experiment of diploid triploid cobitid fishes, Cobitis sinensis-longicorpus complex (Pisces, Cobitidae). Folia Zool 49: 17–22.
Morishima K, Horie S, Yamaha E, Arai K (2002). A cryptic clonal line of the loach Misgurnus anguillicaudatus (Teleostei: Cobitidae) evidenced by induced gynogenesis, interspecific hybridization, microsatellite genotyping and multilocus DNA fingerprinting. Zool Sci 19: 565–575.
Morishima K, Nakamura-Shiokawa Y, Bando E, Li YJ, Boron A, Khan MM et al. (2008a). Cryptic clonal lineages and genetic diversity in the loach Misgurnus anguillicaudatus (Teleostei: Cobitidae) inferred from nuclear and mitochondrial DNA analyses. Genetica 132: 159–171.
Morishima K, Nakayama I, Arai K (2001). Microsatellite-centromere mapping in the loach, Misgurnus anguillicaudatus. Genetica 111: 59–69.
Morishima K, Nakayama I, Arai K (2008b). Genetic linkage map of the loach Misgurnus anguillicaudatus (Teleostei: Cobitidae). Genetica 132: 227–241.
Nishioka M, Ohtani H (1984). Hybridogenetic reproduction of allotriploids between Japanese and European pond frog. Zool Sci 1: 291–316.
Ohtani H (1993). Mechanism of chromosome elimination in the hybridogenetic spermatogenesis of allotriploid males between Japanese and European water frogs. Chromosoma 102: 158–162.
Oshima K, Morishima K, Yamaha E, Arai K (2005). Reproductive capacity of triploid loaches obtained from Hokkaido Island, Japan. Ichthyol Res 52: 1–8.
Saitoh K (1989). Asian pond loach. In: Kawanabe H, Mzuno N (eds). Freshwater fishes of Japan. Yamakei Pub: Tokyo. pp 382–385.
Saitoh K, Kim IS, Lee EH (2004). Mitochondrial gene introgression between spined loaches via hybridogenesis. Zool Sci 21: 795–798.
Schultz RJ (1961). Reproductive mechanisms of unisexual and bisexual strains of viviparous fish Poeciliopsis. Evolution 15: 302–325.
Schultz RJ (1966). Hybridization experiments with an all-female fish of the genus Poeciliopsis. Biol Bull 130: 415–429.
Schultz RJ (1969). Hybridization, unisexuality, and polyploidy in the teleost Poeciliopsis and the other vertebrates. Am Nat 103: 605–619.
Tas IC, Van Dijk PJ (1999). Crosses between sexual and apomictic dandelions (Taraxacum). I. The inheritance of apomixis. Heredity 83: 707–714.
van Dijk PJ, Bakx-Schotman JM (2004). Formation of unreduced megaspores (diplospory) in apomictic dandelions (Taraxacum officinale, s.l.) is controlled by a sex-specific dominant locus. Genetics 166: 483–492.
van Dijk PJ, Tas IC, Falque M, Bakx-Schotman T (1999). Crosses between sexual and apomictic dandelions (Taraxacum). II. The breakdown of apomixis. Heredity 83: 715–721.
Vrijenhoek RC, Dawley RM, Cole CJ, Bogart JP (1989). A list of the known unisexual vertebrates. In: Dawley RM, Bogart T JP (eds). Evolution and Ecology of Unisexual Vertebrates. New York State Museum: Albany. Bulletin 466, pp 19–23.
Yoshikawa H, Morishima K, Kusuda S, Yamaha E, Arai K (2007). Diploid sperm produced by artificially sex-reversed clone loaches. J Exp Zool 307A: 75–83.
Zhang Q, Arai K (1999). Distribution and reproductive capacity of natural triploid individuals and occurrence of unreduced eggs as a cause of polyploidization in the loach, Misgurnus anguillicaudatus. Ichthyol Res 46: 153–161.
Zhang Q, Arai K, Yamashita M (1998). Cytogenetic mechanisms for triploid and haploid eggs formation in the loach, Misgurnus anguillicaudatus. J Exp Zool 281: 608–619.
Acknowledgements
This study was partly supported by Grants-in-Aid for the 21st Century COE (Center of Excellence) Program from the Ministry of Education, Culture, Sports, Science and Technology, Japan and for Scientific Research (B) (no. 18380108) from the Japan Society for the Promotion of Sciences.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Morishima, K., Yoshikawa, H. & Arai, K. Meiotic hybridogenesis in triploid Misgurnus loach derived from a clonal lineage. Heredity 100, 581–586 (2008). https://doi.org/10.1038/hdy.2008.17
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/hdy.2008.17
Keywords
This article is cited by
-
Development of a repetitive DNA marker for identification of the exotic large-scale loach introduced to Japan
Fisheries Science (2024)
-
Meiosis and gametogenesis in hybrid, polyploid, and clonal fishes: case studies in the dojo loach Misgurnus anguillicaudatus
Fisheries Science (2023)
-
Genotypic similarities among the parthenogenetic Darevskia rock lizards with different hybrid origins
BMC Evolutionary Biology (2020)
-
The programmed DNA elimination and formation of micronuclei in germ line cells of the natural hybridogenetic water frog Pelophylax esculentus
Scientific Reports (2018)
-
Development of nuclear DNA markers to characterize genetically diverse groups of Misgurnus anguillicaudatus and its closely related species
Fisheries Science (2017)