Abstract
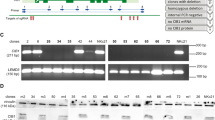
Cutaneous gene therapy can be envisioned through the use of keratinocyte stem cell clones in which retroviral genotoxic risks can be pre-assessed. While transactivation of cellular genes by the retroviral long terminal repeat enhancer has been proven in experimental and clinical settings, the formation of chimeric viral–cellular transcripts originated by the inefficient termination (read-through) of retroviral transcripts remains to be studied in depth. We now demonstrate the widespread presence of viral–cellular fusion transcripts derived from integrated proviruses in keratinocytes transduced with self-inactivating (SIN) retroviral vectors. We have detected high molecular weight RNAs in northern blot analysis of retroviral vector expression in individual cell clones. Characterization of some of these transcripts revealed that they originate from genes located at the proviral integration sites. One class of transcripts corresponds to fusions of the viral vectors with intronic sequences, terminating at cryptic polyadenylation sites located in introns. A second class comprises fusion transcripts with coding sequences of genes at the integration sites. These are generated through splicing from a cryptic, not previously described donor site in the lentiviral vectors to exons of cellular genes, and have the potential to encode unintended open reading frames, although they are downregulated by cellular mechanisms. Our data contribute to a better understanding of the impact of SIN lentiviral vector integration on cellular gene transcription, and will be helpful in improving the design of this type of vectors.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
De Luca M, Pellegrini G, Mavilio F . Gene therapy of inherited skin adhesion disorders: a critical overview. Br J Dermatol 2009; 161: 19–24.
Aiuti A, Roncarolo MG . Ten years of gene therapy for primary immune deficiencies. Hematology Am Soc Hematol Educ Program 2009: 682–689.
Cartier N, Hacein-Bey-Abina S, Bartholomae CC, Veres G, Schmidt M, Kutschera I et al. Hematopoietic stem cell gene therapy with a lentiviral vector in X-linked adrenoleukodystrophy. Science 2009; 326: 818–823.
Larcher F, Dellambra E, Rico L, Bondanza S, Murillas R, Cattoglio C et al. Long-term engraftment of single genetically modified human epidermal holoclones enables safety pre-assessment of cutaneous gene therapy. Mol Ther 2007; 15: 1670–1676.
Modlich U, Navarro S, Zychlinski D, Maetzig T, Knoess S, Brugman MH et al. Insertional transformation of hematopoietic cells by self-inactivating lentiviral and gammaretroviral vectors. Mol Ther 2009; 17: 1919–1928.
Uren AG, Kool J, Berns A, van Lohuizen M . Retroviral insertional mutagenesis: past, present and future. Oncogene 2005; 24: 7656–7672.
Nilsen TW, Maroney PA, Goodwin RG, Rottman FM, Crittenden LB, Raines MA et al. c-erbB activation in ALV-induced erythroblastosis: novel RNA processing and promoter insertion result in expression of an amino-truncated EGF receptor. Cell 1985; 41: 719–726.
Herman SA, Coffin JM . Differential transcription from the long terminal repeats of integrated avian leukosis virus DNA. J Virol 1986; 60: 497–505.
Swain A, Coffin JM . Influence of sequences in the long terminal repeat and flanking cell DNA on polyadenylation of retroviral transcripts. J Virol 1993; 67: 6265–6269.
Zaiss AK, Son S, Chang LJ . RNA 3′ readthrough of oncoretrovirus and lentivirus: implications for vector safety and efficacy. J Virol 2002; 76: 7209–7219.
Hendrie PC, Huo Y, Stolitenko RB, Russell DW . A rapid and quantitative assay for measuring neighboring gene activation by vector proviruses. Mol Ther 2008; 16: 534–540.
Yang Q, Lucas A, Son S, Chang LJ . Overlapping enhancer/promoter and transcriptional termination signals in the lentiviral long terminal repeat. Retrovirology 2007; 4: 4.
Li Z, Dullmann J, Schiedlmeier B, Schmidt M, von Kalle C, Meyer J et al. Murine leukemia induced by retroviral gene marking. Science 2002; 296: 497.
Montini E, Cesana D, Schmidt M, Sanvito F, Bartholomae CC, Ranzani M et al. The genotoxic potential of retroviral vectors is strongly modulated by vector design and integration site selection in a mouse model of HSC gene therapy. J Clin Invest 2009; 119: 964–975.
Di Nunzio F, Maruggi G, Ferrari S, Di Iorio E, Poletti V, Garcia M et al. Correction of laminin-5 deficiency in human epidermal stem cells by transcriptionally targeted lentiviral vectors. Mol Ther 2008; 16: 1977–1985.
Carter MS, Doskow J, Morris P, Li S, Nhim RP, Sandstedt S et al. A regulatory mechanism that detects premature nonsense codons in T-cell receptor transcripts in vivo is reversed by protein synthesis inhibitors in vitro. J Biol Chem 1995; 270: 28995–29003.
Schambach A, Galla M, Maetzig T, Loew R, Baum C . Improving transcriptional termination of self-inactivating gamma-retroviral and lentiviral vectors. Mol Ther 2007; 15: 1167–1173.
Nienhuis AW, Dunbar CE, Sorrentino BP . Genotoxicity of retroviral integration in hematopoietic cells. Mol Ther 2006; 13: 1031–1049.
Zufferey R, Nagy D, Mandel RJ, Naldini L, Trono D . Multiply attenuated lentiviral vector achieves efficient gene delivery in vivo. Nat Biotechnol 1997; 15: 871–875.
Bhuvanagiri M, Schlitter AM, Hentze MW, Kulozik AE . NMD: RNA biology meets human genetic medicine. Biochem J 2010; 430: 365–377.
Calvo SE, Pagliarini DJ, Mootha VK . Upstream open reading frames cause widespread reduction of protein expression and are polymorphic among humans. Proc Natl Acad Sci USA 2009; 106: 7507–7512.
Wang CL, Wang BB, Bartha G, Li L, Channa N, Klinger M et al. Activation of an oncogenic microRNA cistron by provirus integration. Proc Natl Acad Sci USA 2006; 103: 18680–18684.
Wu X, Li Y, Crise B, Burgess SM . Transcription start regions in the human genome are favored targets for MLV integration. Science 2003; 300: 1749–1751.
Cattoglio C, Facchini G, Sartori D, Antonelli A, Miccio A, Cassani B et al. Hot spots of retroviral integration in human CD34+ hematopoietic cells. Blood 2007; 110: 1770–1778.
Acknowledgements
We thank Mrs Blanca Duarte for excellent technical help. This work was supported in part by grants, SAF2007-61019 from the Spanish Ministry of Science and Innovation; PI081054 from FIS and P-BIO-0306-2006 from Comunidad de Madrid. The CIBER for rare diseases is an initiative of the Spanish ISCIII.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies the paper on Gene Therapy website
Rights and permissions
About this article
Cite this article
Almarza, D., Bussadori, G., Navarro, M. et al. Risk assessment in skin gene therapy: viral–cellular fusion transcripts generated by proviral transcriptional read-through in keratinocytes transduced with self-inactivating lentiviral vectors. Gene Ther 18, 674–681 (2011). https://doi.org/10.1038/gt.2011.12
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/gt.2011.12
Keywords
This article is cited by
-
Gene Therapy for SCID
Current Pediatrics Reports (2015)
-
Uncovering and Dissecting the Genotoxicity of Self-inactivating Lentiviral Vectors In Vivo
Molecular Therapy (2014)
-
Long-Term Follow-up of Foamy Viral Vector-Mediated Gene Therapy for Canine Leukocyte Adhesion Deficiency
Molecular Therapy (2013)
-
Lack of genotoxicity due to foamy virus vector integration in human iPSCs
Gene Therapy (2013)
-
Highly Efficient Zinc-Finger Nuclease-Mediated Disruption of an eGFP Transgene in Keratinocyte Stem Cells without Impairment of Stem Cell Properties
Stem Cell Reviews and Reports (2012)